Applications of the class II lanthipeptide protease LicP for sequence-specific, traceless peptide bond cleavage.
Tang, W., Dong, S.H., Repka, L.M., He, C., Nair, S.K., van der Donk, W.A.(2015) Chem Sci 6: 6270-6279
- PubMed: 30090246
- DOI: https://doi.org/10.1039/c5sc02329g
- Primary Citation of Related Structures:
4ZOQ - PubMed Abstract:
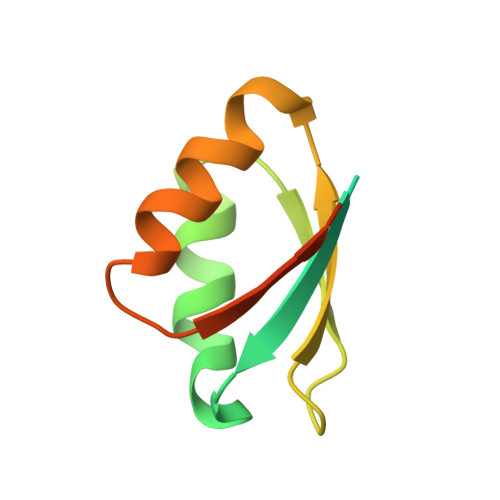
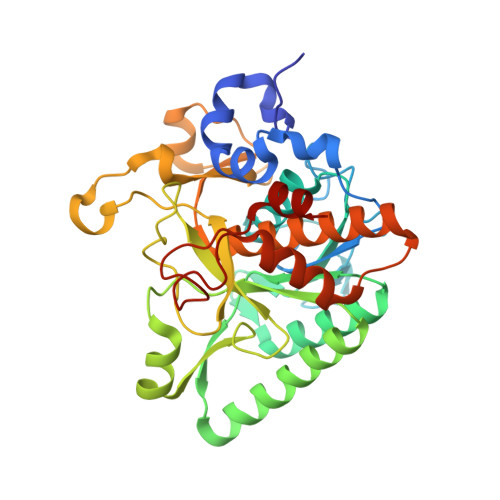
The final step of lanthipeptide biosynthesis involves the removal of leader peptides by dedicated proteases. In vitro characterization of LicP, a class II LanP protease involved in the biosynthesis of the lantibiotic lichenicidin, revealed a self-cleavage step that removes 100 amino acids from the N-terminus. The 2.35 Å resolution crystal structure provides insights into the active site geometry and substrate specificity, and unveiled an unusual calcium-independent maturation mechanism of a subtilisin family member. LicP processes LicA2 peptides with or without post-translational modifications, but dehydrated and cyclized LicA2 is favored. Investigation of its substrate specificity demonstrated that LicP can serve as an efficient sequence-specific traceless protease and may have great utility in basic research and biotechnology. Encouraged by these findings for LicP, we identified 13 other class II LanPs, ten of which were previously unknown, and suggest that these proteins may serve as a pool of proteases with diverse recognition sequences for general traceless tag removal applications, expanding the current toolbox of proteases.
- Department of Chemistry and Howard Hughes Medical Institute , University of Illinois at Urbana-Champaign , 600 S. Mathews Ave. , Urbana , IL 61801 , USA . Email: vddonk@illinois.edu ; ; Tel: +1 217 244 5360.
Organizational Affiliation: