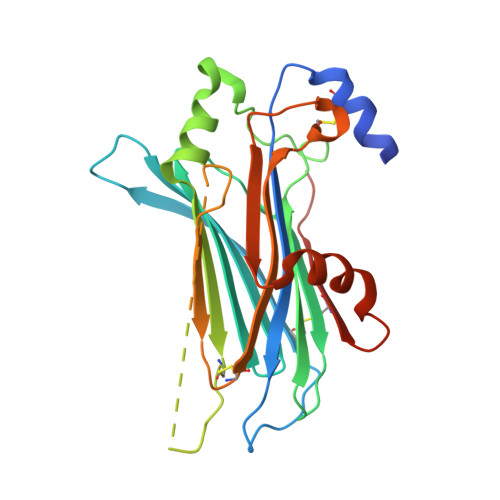
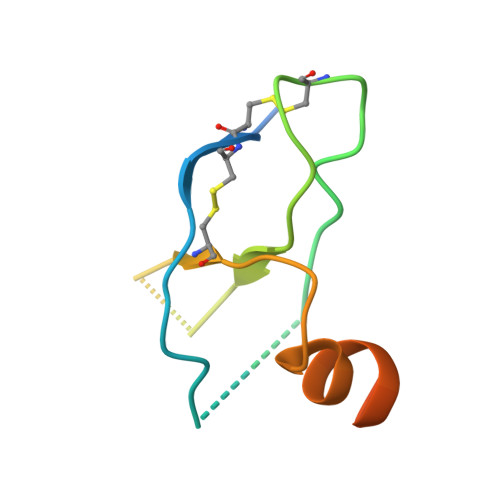
Structures of Orf Virus Chemokine Binding Protein in Complex with Host Chemokines Reveal Clues to Broad Binding Specificity.
Counago, R.M., Knapp, K.M., Nakatani, Y., Fleming, S.B., Corbett, M., Wise, L.M., Mercer, A.A., Krause, K.L.(2015) Structure 23: 1199-1213
- PubMed: 26095031
- DOI: https://doi.org/10.1016/j.str.2015.04.023
- Primary Citation of Related Structures:
4P5I, 4ZK9, 4ZKB, 4ZKC - PubMed Abstract:
The chemokine binding protein (CKBP) from orf virus (ORFV) binds with high affinity to chemokines from three classes, C, CC, and CXC, making it unique among poxvirus CKBPs described to date. We present its crystal structure alone and in complex with three CC chemokines, CCL2, CCL3, and CCL7. ORFV CKBP possesses a β-sandwich fold that is electrostatically and sterically complementary to its binding partners. Chemokines bind primarily through interactions involving the N-terminal loop and a hydrophobic recess on the ORFV CKBP β-sheet II surface, and largely polar interactions between the chemokine 20s loop and a negatively charged surface groove located at one end of the CKBP β-sheet II surface. ORFV CKBP interacts with leukocyte receptor and glycosaminoglycan binding sites found on the surface of bound chemokines. SEC-MALLS and chromatographic evidence is presented supporting that ORFV CKBP is a dimer in solution over a broad range of protein concentrations.
- Department of Biochemistry, University of Otago, Dunedin 9054, New Zealand.
Organizational Affiliation: