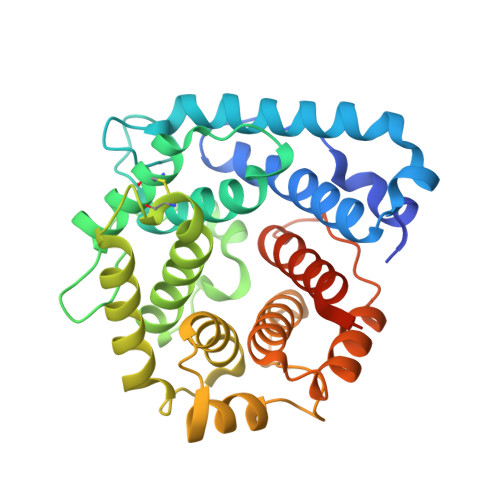
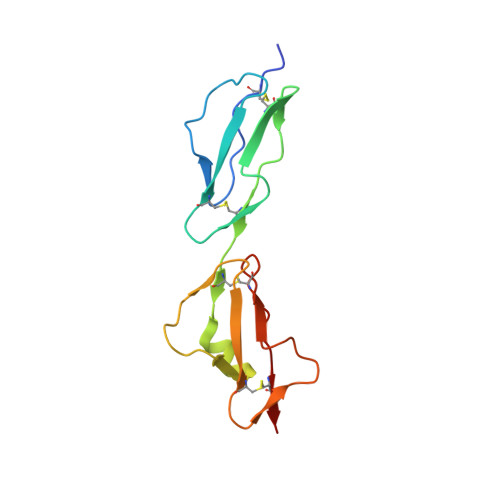
Complement Factor H and Simian Virus 40 bind the GM1 ganglioside in distinct conformations.
Blaum, B.S., Frank, M., Walker, R.C., Neu, U., Stehle, T.(2016) Glycobiology 26: 532-539
- PubMed: 26715202
- DOI: https://doi.org/10.1093/glycob/cwv170
- Primary Citation of Related Structures:
4ZH1 - PubMed Abstract:
Mammalian cell surfaces are decorated with a variety of glycan chains that orchestrate development and defense and are exploited by pathogens for cellular attachment and entry. While glycosidic linkages are, in principle, flexible, the conformational space that a given glycan can sample is subject to spatial and electrostatic restrictions imposed by its overall chemical structure. Here, we show how the glycan moiety of the GM1 ganglioside, a branched, monosialylated pentasaccharide that serves as a ligand for various proteins, undergoes differential conformational selection in its interactions with different lectins. Using STD NMR and X-ray crystallography, we found that the innate immune regulator complement Factor H (FH) binds a previously not reported GM1 conformation that is not compatible with the GM1-binding sites of other structurally characterized GM1-binding lectins such as the Simian Virus 40 (SV40) capsid. Molecular dynamics simulations of the free glycan in explicit solvent on the 10 μs timescale reveal that the FH-bound conformation nevertheless corresponds to a minimum in the Gibbs free energy plot. In contrast to the GM1 conformation recognized by SV40, the FH-bound GM1 conformation is associated with poor NOE restraints, explaining how it escaped(1)H-(1)H NOE-restrained modeling in the past and highlighting the necessity for ensemble representations of glycan structures.
- Interfaculty Institute of Biochemistry, University of Tübingen, Tübingen 72076, Germany baerbel.blaum@uni-tuebingen.de.
Organizational Affiliation: