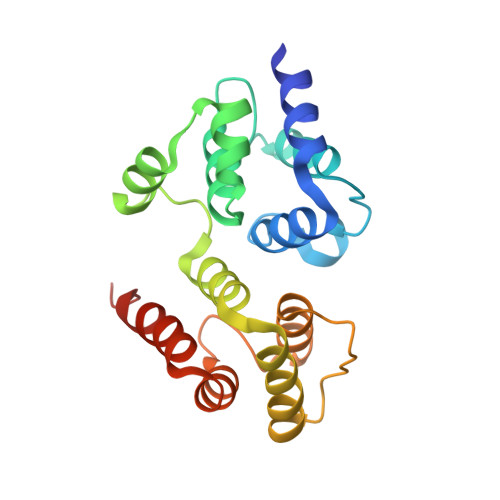
Crystal structure of the death effector domains of caspase-8
Shen, C., Yue, H., Pei, J., Guo, X., Wang, T., Quan, J.M.(2015) Biochem Biophys Res Commun 463: 297-302
- PubMed: 26003730
- DOI: https://doi.org/10.1016/j.bbrc.2015.05.054
- Primary Citation of Related Structures:
4ZBW - PubMed Abstract:
Caspase-8 is a key mediator in various biological processes such as apoptosis, necroptosis, inflammation, T/B cells activation, and cell motility. Caspase-8 is characterized by the N-terminal tandem death effector domains (DEDs) and the C-terminal catalytic protease domain. The DEDs mediate diverse functions of caspase-8 through homotypic interactions of the DEDs between caspase-8 and its partner proteins. Here, we report the first crystal structure of the DEDs of caspase-8. The overall structure of the DEDs of caspase-8 is similar to that of the DEDs of vFLIP MC159, which is composed of two tandem death effector domains that closely associate with each other in a head-to-tail manner. Structural analysis reveals distinct differences in the region connecting helices α2b and α4b in the second DED of the DEDs between caspase-8 and MC159, in which the helix α3b in MC159 is replaced by a loop in caspase-8. Moreover, the different amino acids in this region might confer the distinct features of solubility and aggregation for the DEDs of caspase-8 and MC159.
- Key Laboratory of Structural Biology, School of Chemical Biology & Biotechnology, Peking University, Shenzhen Graduate School, Shenzhen 518055, China.
Organizational Affiliation: