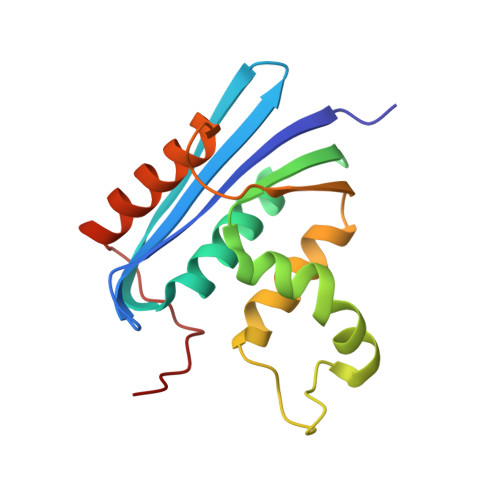
Interaction with Single-stranded DNA-binding Protein Stimulates Escherichia coli Ribonuclease HI Enzymatic Activity.
Petzold, C., Marceau, A.H., Miller, K.H., Marqusee, S., Keck, J.L.(2015) J Biological Chem 290: 14626-14636
- PubMed: 25903123
- DOI: https://doi.org/10.1074/jbc.M115.655134
- Primary Citation of Related Structures:
4Z0U - PubMed Abstract:
Single-stranded (ss) DNA-binding proteins (SSBs) bind and protect ssDNA intermediates formed during replication, recombination, and repair reactions. SSBs also directly interact with many different genome maintenance proteins to stimulate their enzymatic activities and/or mediate their proper cellular localization. We have identified an interaction formed between Escherichia coli SSB and ribonuclease HI (RNase HI), an enzyme that hydrolyzes RNA in RNA/DNA hybrids. The RNase HI·SSB complex forms by RNase HI binding the intrinsically disordered C terminus of SSB (SSB-Ct), a mode of interaction that is shared among all SSB interaction partners examined to date. Residues that comprise the SSB-Ct binding site are conserved among bacterial RNase HI enzymes, suggesting that RNase HI·SSB complexes are present in many bacterial species and that retaining the interaction is important for its cellular function. A steady-state kinetic analysis shows that interaction with SSB stimulates RNase HI activity by lowering the reaction Km. SSB or RNase HI protein variants that disrupt complex formation nullify this effect. Collectively our findings identify a direct RNase HI/SSB interaction that could play a role in targeting RNase HI activity to RNA/DNA hybrid substrates within the genome.
- From the Department of Biomolecular Chemistry, University of Wisconsin School of Medicine and Public Health, Madison, Wisconsin 53706 and.
Organizational Affiliation: