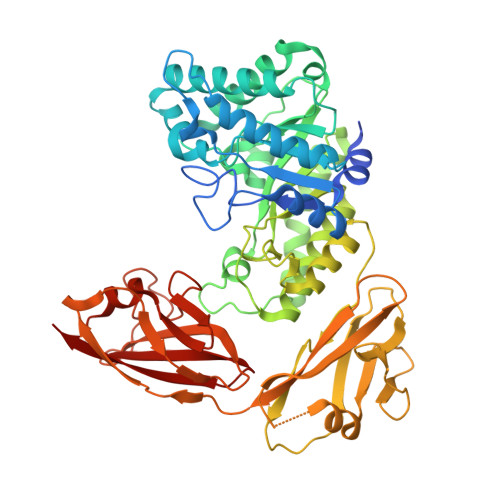
Molecular characterization of a family 5 glycoside hydrolase suggests an induced-fit enzymatic mechanism.
Liberato, M.V., Silveira, R.L., Prates, E.T., de Araujo, E.A., Pellegrini, V.O., Camilo, C.M., Kadowaki, M.A., de O.Neto, M., Popov, A., Skaf, M.S., Polikarpov, I.(2016) Sci Rep 6: 23473-23473
- PubMed: 27032335
- DOI: https://doi.org/10.1038/srep23473
- Primary Citation of Related Structures:
4YZP, 4YZT - PubMed Abstract:
Glycoside hydrolases (GHs) play fundamental roles in the decomposition of lignocellulosic biomaterials. Here, we report the full-length structure of a cellulase from Bacillus licheniformis (BlCel5B), a member of the GH5 subfamily 4 that is entirely dependent on its two ancillary modules (Ig-like module and CBM46) for catalytic activity. Using X-ray crystallography, small-angle X-ray scattering and molecular dynamics simulations, we propose that the C-terminal CBM46 caps the distal N-terminal catalytic domain (CD) to establish a fully functional active site via a combination of large-scale multidomain conformational selection and induced-fit mechanisms. The Ig-like module is pivoting the packing and unpacking motions of CBM46 relative to CD in the assembly of the binding subsite. This is the first example of a multidomain GH relying on large amplitude motions of the CBM46 for assembly of the catalytically competent form of the enzyme.
- São Carlos Institute of Physics, University of São Paulo, São Carlos 13566-590 São Paulo, Brazil.
Organizational Affiliation: