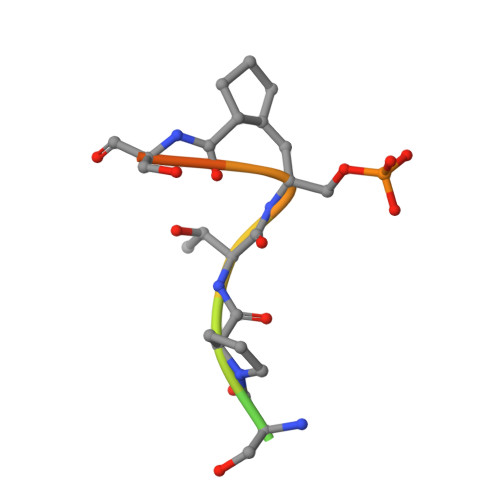
Chemical Tools To Decipher Regulation of Phosphatases by Proline Isomerization on Eukaryotic RNA Polymerase II.
Mayfield, J.E., Fan, S., Wei, S., Zhang, M., Li, B., Ellington, A.D., Etzkorn, F.A., Zhang, Y.J.(2015) ACS Chem Biol 10: 2405-2414
- PubMed: 26332362
- DOI: https://doi.org/10.1021/acschembio.5b00296
- Primary Citation of Related Structures:
4YGX, 4YGY, 4YH1 - PubMed Abstract:
Proline isomerization greatly impacts biological signaling but is subtle and difficult to detect in proteins. We characterize this poorly understood regulatory mechanism for RNA polymerase II carboxyl terminal domain (CTD) phosphorylation state using novel, direct, and quantitative chemical tools. We determine the proline isomeric preference of three CTD phosphatases: Ssu72 as cis-proline specific, Scp1 and Fcp1 as strongly trans-preferred. Due to this inherent characteristic, these phosphatases respond differently to enzymes that catalyze the isomerization of proline, like Ess1/Pin1. We demonstrate that this selective regulation of RNA polymerase II phosphorylation state exists within human cells, consistent with in vitro assays. These results support a model in which, instead of a global enhancement of downstream enzymatic activities, proline isomerases selectively boost the activity of a subset of CTD regulatory factors specific for cis-proline. This leads to diversified phosphorylation states of CTD in vitro and in cells. We provide the chemical tools to investigate proline isomerization and its ability to selectively enhance signaling in transcription and other biological contexts.
- Department of Molecular Biosciences and Institute for Cellular and Molecular Biology, University of Texas at Austin , Austin, Texas 78712, United States.
Organizational Affiliation: