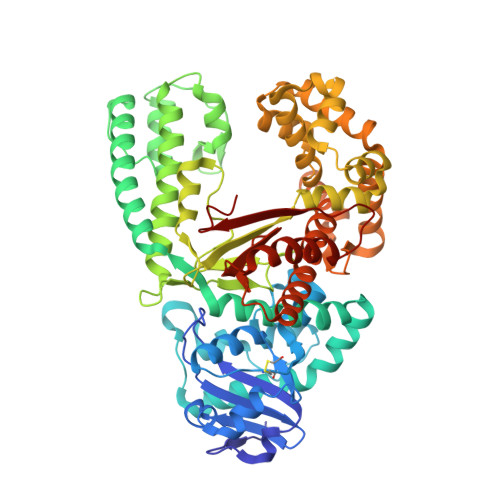
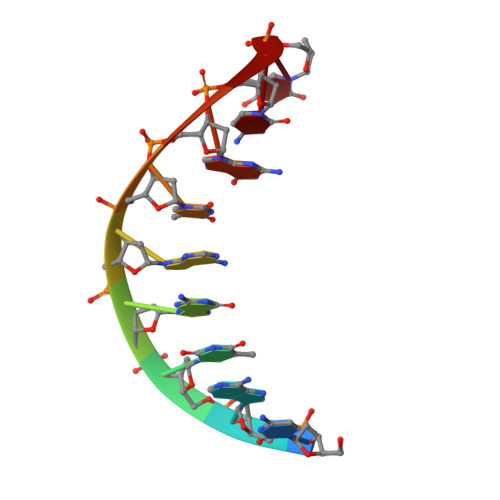
The Closing Mechanism of DNA Polymerase I at Atomic Resolution.
Miller, B.R., Beese, L.S., Parish, C.A., Wu, E.Y.(2015) Structure 23: 1609-1620
- PubMed: 26211612
- DOI: https://doi.org/10.1016/j.str.2015.06.016
- Primary Citation of Related Structures:
4YFU - PubMed Abstract:
DNA polymerases must quickly and accurately distinguish between similar nucleic acids to form Watson-Crick base pairs and avoid DNA replication errors. Deoxynucleoside triphosphate (dNTP) binding to the DNA polymerase active site induces a large conformational change that is difficult to characterize experimentally on an atomic level. Here, we report an X-ray crystal structure of DNA polymerase I bound to DNA in the open conformation with a dNTP present in the active site. We use this structure to computationally simulate the open to closed transition of DNA polymerase in the presence of a Watson-Crick base pair. Our microsecond simulations allowed us to characterize the key steps involved in active site assembly, and propose the sequence of events involved in the prechemistry steps of DNA polymerase catalysis. They also reveal new features of the polymerase mechanism, such as a conserved histidine as a potential proton acceptor from the primer 3'-hydroxyl.
- Department of Biology, University of Richmond, 28 Westhampton Way, Richmond, VA 23173, USA; Department of Chemistry, University of Richmond, 28 Westhampton Way, Richmond, VA 23173, USA.
Organizational Affiliation: