Structure of Acinetobacter baumannii bacteriophage AP22 polysaccharide degrading lyase in complex with A. baumannii capsular saccharide at 0.9 A resolution
Buth, S.A., Shneider, M.M., Leiman, P.G.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Particle-associated lyase | 625 | Acinetobacter phage AP22 | Mutation(s): 0 | 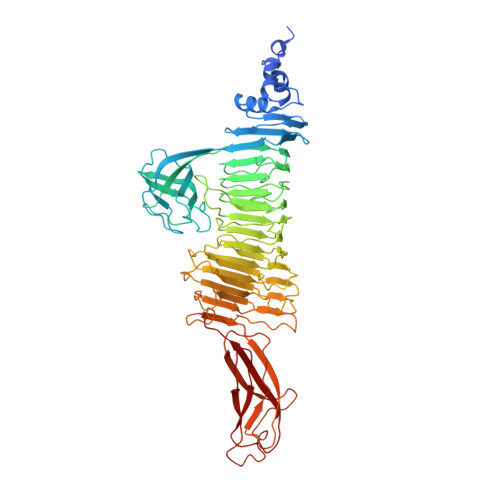 | |
UniProt | |||||
Find proteins for I2GUG1 (Acinetobacter phage AP22) Explore I2GUG1 Go to UniProtKB: I2GUG1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | I2GUG1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| 2-acetamido-2,4-dideoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-alpha-D-fucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-mannopyranuronic acid | B | 3 |  | N/A | |
Glycosylation Resources | |||||
GlyTouCan: G54331LR GlyCosmos: G54331LR | |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SO4 Query on SO4 | K [auth A], L [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| EDO Query on EDO | C [auth A] D [auth A] E [auth A] F [auth A] G [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| CL Query on CL | M [auth A], O [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| NA Query on NA | N [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 92.634 | α = 90 |
| b = 92.634 | β = 90 |
| c = 391.478 | γ = 120 |
| Software Name | Purpose |
|---|---|
| Coot | model building |
| XDS | data reduction |
| XSCALE | data scaling |
| SHELX | phasing |
| SHELXL | refinement |
| SHELXL-97 | refinement |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Ecole Polytechnique Federale de Lausanne | Switzerland | 0577-1 |