Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
Huber, E.M., de Bruin, G., Heinemeyer, W., Paniagua Soriano, G., Overkleeft, H.S., Groll, M.(2015) J Am Chem Soc 137: 7835-7842
- PubMed: 26020686
- DOI: https://doi.org/10.1021/jacs.5b03688
- Primary Citation of Related Structures:
4Y69, 4Y6A, 4Y6V, 4Y6Z, 4Y70, 4Y74, 4Y75, 4Y77, 4Y78, 4Y7W, 4Y7X, 4Y7Y, 4Y80, 4Y81, 4Y82, 4Y84, 4Y8G, 4Y8H, 4Y8I, 4Y8J, 4Y8K, 4Y8L, 4Y8M, 4Y8N, 4Y8O, 4Y8P, 4Y8Q, 4Y8R, 4Y8S, 4Y8T, 4Y8U, 4Y9Y, 4Y9Z, 4YA0, 4YA1, 4YA2, 4YA3, 4YA4, 4YA5, 4YA7, 4YA9 - PubMed Abstract:
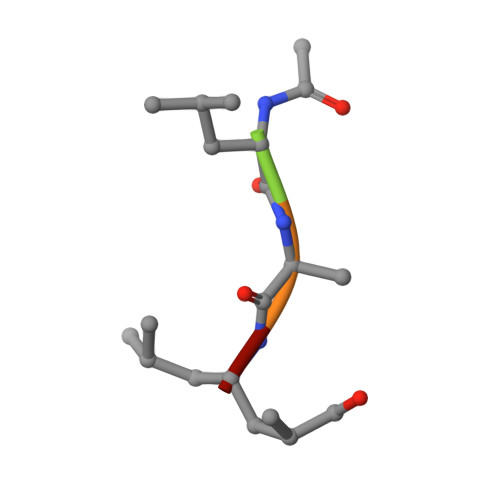
Cleavage analyses of 20S proteasomes with natural or synthetic substrates allowed to infer the substrate specificities of the active sites and paved the way for the rational design of high-affinity proteasome inhibitors. However, details of cleavage preferences remained enigmatic due to the lack of appropriate structural data. In a unique approach, we here systematically examined substrate specificities of yeast and human proteasomes using irreversibly acting α',β'epoxyketone (ep) inhibitors. Biochemical and structural analyses provide unique insights into the substrate preferences of the distinct active sites and highlight differences between proteasome types that may be considered in future inhibitor design efforts. (1) For steric reasons, epoxyketones with Val or Ile at the P1 position are weak inhibitors of all active sites. (2) Identification of the β2c selective compound Ac-LAE-ep represents a promising starting point for the development of compounds that discriminate between β2c and β2i. (3) The compound Ac-LAA-ep was found to favor subunit β5c over β5i by three orders of magnitude. (4) Yeast β1 and human β1c subunits preferentially bind Asp and Leu in their S1 pockets, while Glu and large hydrophobic residues are not accepted. (5) Exceptional structural features in the β1/2 substrate binding channel give rise to the β1 selectivity of compounds featuring Pro at the P3 site. Altogether, 23 different epoxyketone inhibitors, five proteasome mutants, and 43 crystal structures served to delineate a detailed picture of the substrate and ligand specificities of proteasomes and will further guide drug development efforts toward subunit-specific proteasome inhibitors for applications as diverse as cancer and autoimmune disorders.
- †Center for Integrated Protein Science at the Department Chemie, Lehrstuhl für Biochemie, Technische Universität München, 85748 Garching, Germany.
Organizational Affiliation: