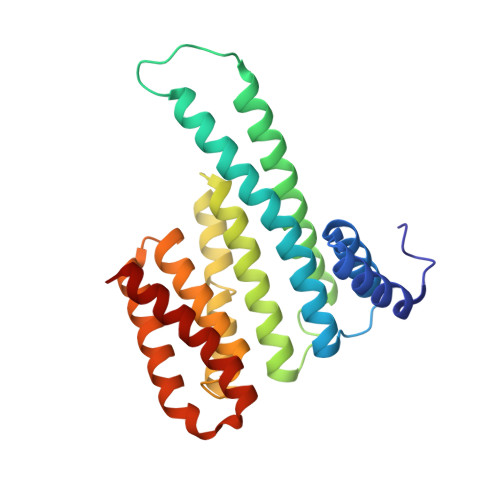
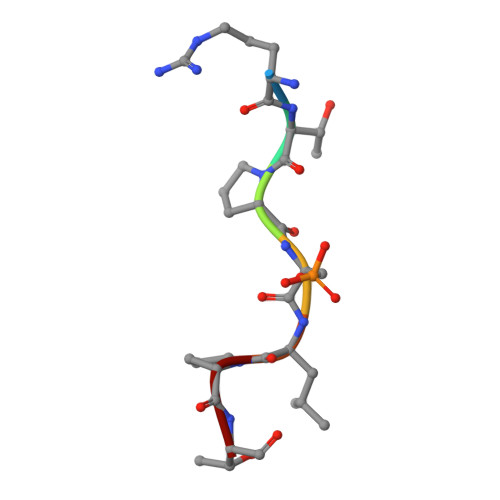
Stabilizer-Guided Inhibition of Protein-Protein Interactions.
Milroy, L.G., Bartel, M., Henen, M.A., Leysen, S., Adriaans, J.M., Brunsveld, L., Landrieu, I., Ottmann, C.(2015) Angew Chem Int Ed Engl 54: 15720-15724
- PubMed: 26537010
- DOI: https://doi.org/10.1002/anie.201507976
- Primary Citation of Related Structures:
4Y32, 4Y5I, 5HF3 - PubMed Abstract:
The discovery of novel protein-protein interaction (PPI) modulators represents one of the great molecular challenges of the modern era. PPIs can be modulated by either inhibitor or stabilizer compounds, which target different though proximal regions of the protein interface. In principle, protein-stabilizer complexes can guide the design of PPI inhibitors (and vice versa). In the present work, we combine X-ray crystallographic data from both stabilizer and inhibitor co-crystal complexes of the adapter protein 14-3-3 to characterize, down to the atomic scale, inhibitors of the 14-3-3/Tau PPI, a potential drug target to treat Alzheimer's disease. The most potent compound notably inhibited the binding of phosphorylated full-length Tau to 14-3-3 according to NMR spectroscopy studies. Our work sets a precedent for the rational design of PPI inhibitors guided by PPI stabilizer-protein complexes while potentially enabling access to new synthetically tractable stabilizers of 14-3-3 and other PPIs.
- Laboratory of Chemical Biology and Institute of Complex Molecular Systems (ICMS), Department of Biomedical Engineering, Technische Universiteit Eindhoven, Den Dolech 2, 5612 AZ Eindhoven (The Netherlands). l.milroy@tue.nl.
Organizational Affiliation: