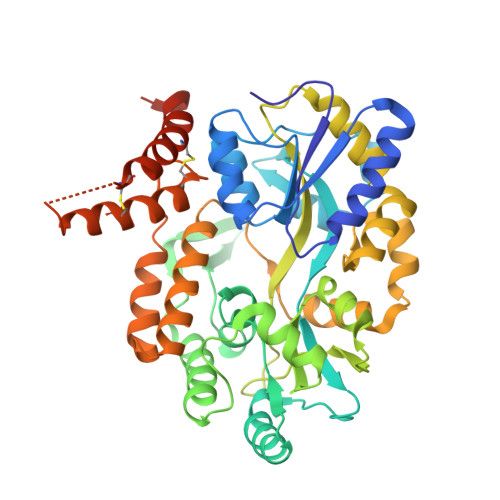
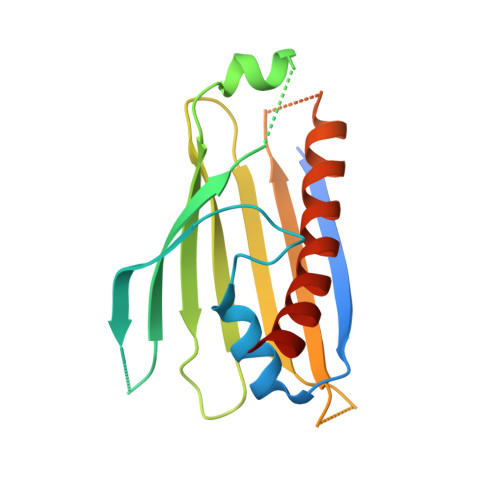
Structural insight into the TRIAP1/PRELI-like domain family of mitochondrial phospholipid transfer complexes.
Miliara, X., Garnett, J.A., Tatsuta, T., Abid Ali, F., Baldie, H., Perez-Dorado, I., Simpson, P., Yague, E., Langer, T., Matthews, S.(2015) EMBO Rep 16: 824-835
- PubMed: 26071602
- DOI: https://doi.org/10.15252/embr.201540229
- Primary Citation of Related Structures:
4XZS, 4XZV - PubMed Abstract:
The composition of the mitochondrial membrane is important for its architecture and proper function. Mitochondria depend on a tightly regulated supply of phospholipid via intra-mitochondrial synthesis and by direct import from the endoplasmic reticulum. The Ups1/PRELI-like family together with its mitochondrial chaperones (TRIAP1/Mdm35) represent a unique heterodimeric lipid transfer system that is evolutionary conserved from yeast to man. Work presented here provides new atomic resolution insight into the function of a human member of this system. Crystal structures of free TRIAP1 and the TRIAP1-SLMO1 complex reveal how the PRELI domain is chaperoned during import into the intermembrane mitochondrial space. The structural resemblance of PRELI-like domain of SLMO1 with that of mammalian phoshatidylinositol transfer proteins (PITPs) suggest that they share similar lipid transfer mechanisms, in which access to a buried phospholipid-binding cavity is regulated by conformationally adaptable loops.
- Department of Life Sciences, Imperial College London, London, UK.
Organizational Affiliation: