Crystal structure of CalS8 from Micromonospora echinospora
Michalska, K., Bigelow, L., Endres, M., Babnigg, G., Bingman, C.A., Yennamalli, R.M., Singh, S., Kharel, M.K., Thorson, J.S., Phillips Jr., G.N., Joachimiak, A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| CalS8 | 456 | Micromonospora echinospora | Mutation(s): 1 Gene Names: calS8 | 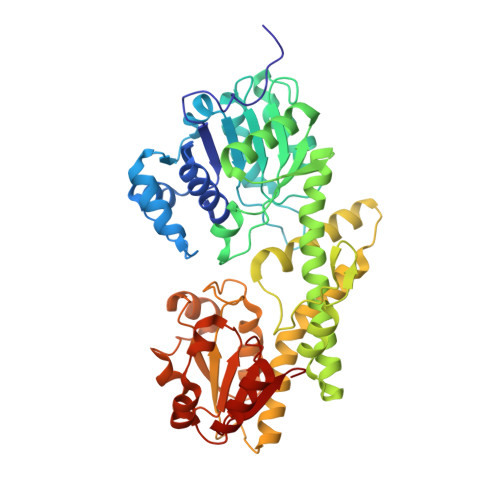 | |
UniProt | |||||
Find proteins for Q8KNF6 (Micromonospora echinospora) Explore Q8KNF6 Go to UniProtKB: Q8KNF6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8KNF6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAD Query on NAD | C [auth A] | NICOTINAMIDE-ADENINE-DINUCLEOTIDE C21 H27 N7 O14 P2 BAWFJGJZGIEFAR-NNYOXOHSSA-N |  | ||
| TMP Query on TMP | E [auth A] | THYMIDINE-5'-PHOSPHATE C10 H15 N2 O8 P GYOZYWVXFNDGLU-XLPZGREQSA-N |  | ||
| GOL Query on GOL | F [auth B] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| NA Query on NA | D [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 70.682 | α = 90 |
| b = 87.04 | β = 103.59 |
| c = 70.66 | γ = 90 |
| Software Name | Purpose |
|---|---|
| BUSTER | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM094585 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM098248 |