A tail of two sites: a bipartite mechanism for recognition of notch ligands by mind bomb e3 ligases.
McMillan, B.J., Schnute, B., Ohlenhard, N., Zimmerman, B., Miles, L., Beglova, N., Klein, T., Blacklow, S.C.(2015) Mol Cell 57: 912-924
- PubMed: 25747658
- DOI: https://doi.org/10.1016/j.molcel.2015.01.019
- Primary Citation of Related Structures:
4TSE, 4XI6, 4XI7, 4XIB - PubMed Abstract:
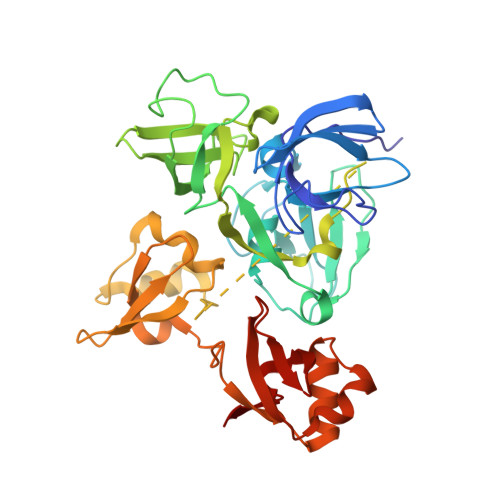
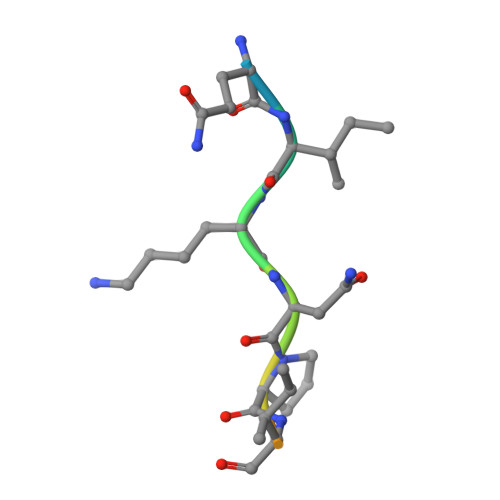
Mind bomb (Mib) proteins are large, multi-domain E3 ligases that promote ubiquitination of the cytoplasmic tails of Notch ligands. This ubiquitination step marks the ligand proteins for epsin-dependent endocytosis, which is critical for in vivo Notch receptor activation. We present here crystal structures of the substrate recognition domains of Mib1, both in isolation and in complex with peptides derived from Notch ligands. The structures, in combination with biochemical, cellular, and in vivo assays, show that Mib1 contains two independent substrate recognition domains that engage two distinct epitopes from the cytoplasmic tail of the ligand Jagged1, one in the intracellular membrane proximal region and the other near the C terminus. Together, these studies provide insights into the mechanism of ubiquitin transfer by Mind bomb E3 ligases, illuminate a key event in ligand-induced activation of Notch receptors, and identify a potential target for therapeutic modulation of Notch signal transduction in disease.
- Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: