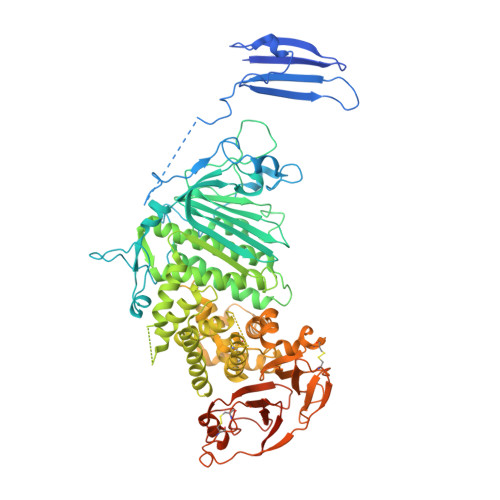
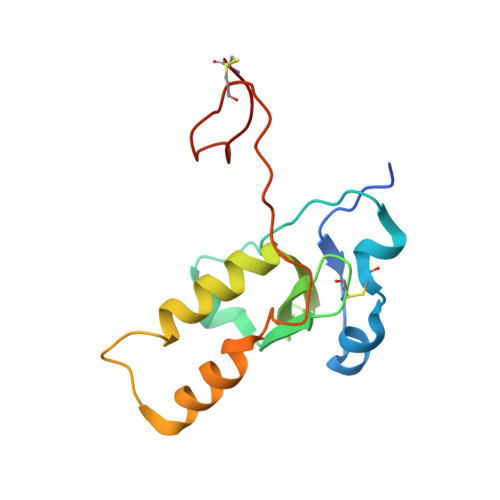
A site of varicella-zoster virus vulnerability identified by structural studies of neutralizing antibodies bound to the glycoprotein complex gHgL.
Xing, Y., Oliver, S.L., Nguyen, T., Ciferri, C., Nandi, A., Hickman, J., Giovani, C., Yang, E., Palladino, G., Grose, C., Uematsu, Y., Lilja, A.E., Arvin, A.M., Carfi, A.(2015) Proc Natl Acad Sci U S A 112: 6056-6061
- PubMed: 25918416
- DOI: https://doi.org/10.1073/pnas.1501176112
- Primary Citation of Related Structures:
4XHJ, 4XI5 - PubMed Abstract:
Varicella-zoster virus (VZV), of the family Alphaherpesvirinae, causes varicella in children and young adults, potentially leading to herpes zoster later in life on reactivation from latency. The conserved herpesvirus glycoprotein gB and the heterodimer gHgL mediate virion envelope fusion with cell membranes during virus entry. Naturally occurring neutralizing antibodies against herpesviruses target these entry proteins. To determine the molecular basis for VZV neutralization, crystal structures of gHgL were determined in complex with fragments of antigen binding (Fabs) from two human monoclonal antibodies, IgG-94 and IgG-RC, isolated from seropositive subjects. These structures reveal that the antibodies target the same site, composed of residues from both gH and gL, distinct from two other neutralizing epitopes identified by negative-stain electron microscopy and mutational analysis. Inhibition of gB/gHgL-mediated membrane fusion and structural comparisons with herpesvirus homologs suggest that the IgG-RC/94 epitope is in proximity to the site on VZV gHgL that activates gB. Immunization studies proved that the anti-gHgL IgG-RC/94 epitope is a critical target for antibodies that neutralize VZV. Thus, the gHgL/Fab structures delineate a site of herpesvirus vulnerability targeted by natural immunity.
- GlaxoSmithKline (GSK) Vaccines, Cambridge, MA 02139; andrea.x.carfi@gsk.com yi.x.xing@gsk.com.
Organizational Affiliation: