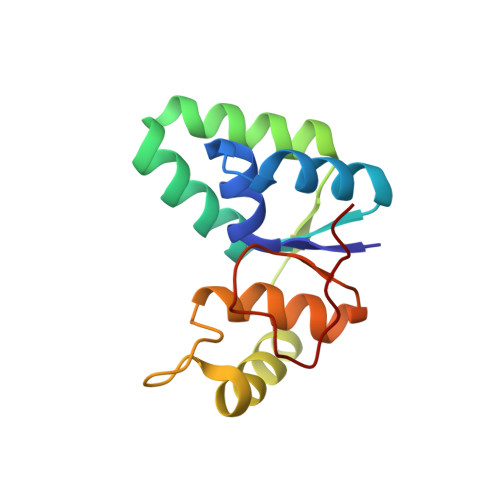
Structural and functional studies of the Mycobacterium tuberculosis VapBC30 toxin-antitoxin system: implications for the design of novel antimicrobial peptides
Lee, I.-G., Lee, S.J., Chae, S., Lee, K.Y., Kim, J.H., Lee, B.-J.(2015) Nucleic Acids Res 43: 7624-7637
- PubMed: 26150422
- DOI: https://doi.org/10.1093/nar/gkv689
- Primary Citation of Related Structures:
4XGQ, 4XGR - PubMed Abstract:
Toxin-antitoxin (TA) systems play important roles in bacterial physiology, such as multidrug tolerance, biofilm formation, and arrest of cellular growth under stress conditions. To develop novel antimicrobial agents against tuberculosis, we focused on VapBC systems, which encompass more than half of TA systems in Mycobacterium tuberculosis. Here, we report that theMycobacterium tuberculosis VapC30 toxin regulates cellular growth through both magnesium and manganese ion-dependent ribonuclease activity and is inhibited by the cognate VapB30 antitoxin. We also determined the 2.7-Å resolution crystal structure of the M. tuberculosis VapBC30 complex, which revealed a novel process of inactivation of the VapC30 toxin via swapped blocking by the VapB30 antitoxin. Our study on M. tuberculosis VapBC30 leads us to design two kinds of VapB30 and VapC30-based novel peptides which successfully disrupt the toxin-antitoxin complex and thus activate the ribonuclease activity of the VapC30 toxin. Our discovery herein possibly paves the way to treat tuberculosis for next generation.
- Research Institute of Pharmaceutical Sciences, College of Pharmacy, Seoul National University, Seoul 151-742, Korea.
Organizational Affiliation: