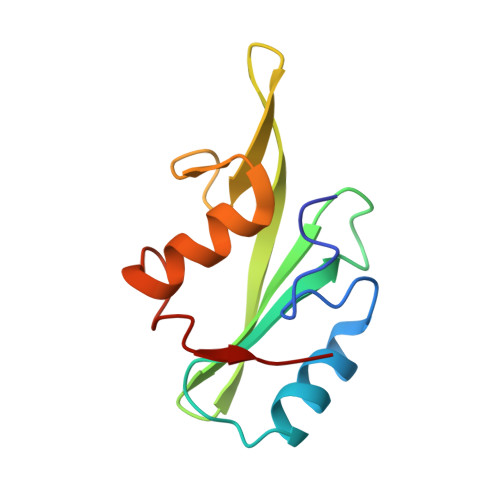
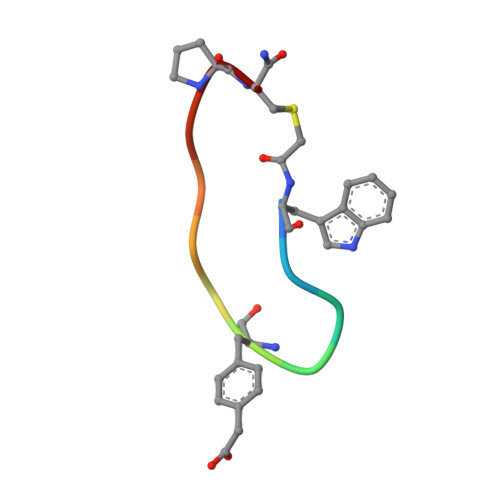
Cyclic Peptides Incorporating Phosphotyrosine Mimetics as Potent and Specific Inhibitors of the Grb7 Breast Cancer Target.
Watson, G.M., Gunzburg, M.J., Ambaye, N.D., Lucas, W.A., Traore, D.A., Kulkarni, K., Cergol, K.M., Payne, R.J., Panjikar, S., Pero, S.C., Perlmutter, P., Wilce, M.C., Wilce, J.A.(2015) J Med Chem 58: 7707-7718
- PubMed: 26359549
- DOI: https://doi.org/10.1021/acs.jmedchem.5b00609
- Primary Citation of Related Structures:
4WWQ, 4X6S - PubMed Abstract:
The Grb7 adaptor protein is a therapeutic target for both TNBC and HER2+ breast cancer. A nonphosphorylated cyclic peptide 1 (known as G7-18NATE) inhibits Grb7 via targeting the Grb7-SH2 domain, but requires the presence of phosphate ions for both affinity and specificity. Here we report the discovery of malonate bound in the phosphotyrosine binding pocket of the apo-Grb7-SH2 structure. Based on this, we carried out the rational design and synthesis of two analogues of peptide 1 that incorporate carboxymethylphenylalanine (cmF) and carboxyphenylalanine (cF) as mimics of phosphotyrosine (pY). Binding studies using SPR confirmed that affinity for Grb7-SH2 domain is improved up to 9-fold over peptide 1 under physiological phosphate conditions (KD = 2.1-5.7 μM) and that binding is specific for Grb7-SH2 over closely related domains (low or no detectable binding to Grb2-SH2 and Grb10-SH2). X-ray crystallographic structural analysis of the analogue bearing a cmF moiety in complex with Grb7-SH2 has identified the precise contacts conferred by the pY mimic that underpin this improved molecular interaction. Together this study identifies and characterizes the tightest specific inhibitor of Grb7 to date, representing a significant development toward a new Grb7-targeted therapeutic.
- School of Chemistry, The University of Sydney , Sydney, New South Wales 2006, Australia.
Organizational Affiliation: