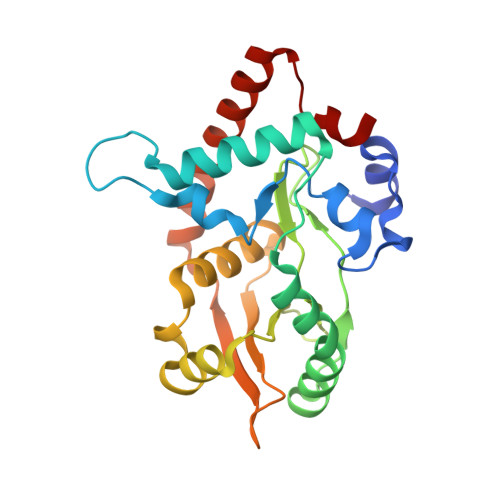
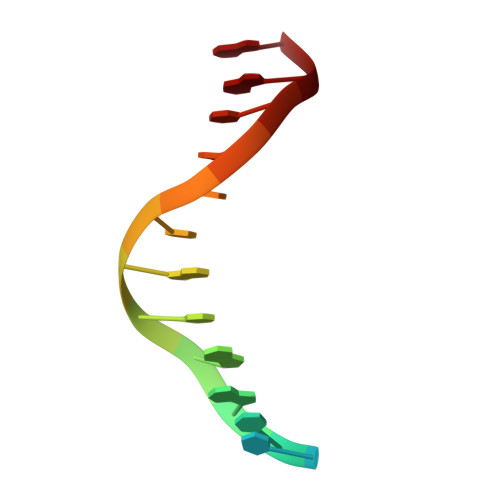
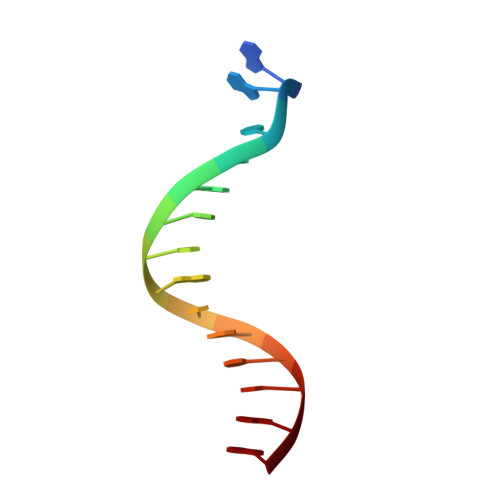
Crystal Structure of lambda Exonuclease in Complex with DNA and Ca(2+).
Zhang, J., Pan, X., Bell, C.E.(2014) Biochemistry 53: 7415-7425
- PubMed: 25370446
- DOI: https://doi.org/10.1021/bi501155q
- Primary Citation of Related Structures:
4WUZ - PubMed Abstract:
Bacteriophage λ exonuclease (λexo) is a ring-shaped homotrimer that resects double-stranded DNA ends in the 5'-3' direction to generate a long 3'-overhang that is a substrate for recombination. λexo is a member of the type II restriction endonuclease-like superfamily of proteins that use a Mg(2+)-dependent mechanism for nucleotide cleavage. A previous structure of λexo in complex with DNA and Mg(2+) was determined using a nuclease defective K131A variant to trap a stable complex. This structure revealed the detailed coordination of the two active site Mg(2+) ions but did not show the interactions involving the side chain of the conserved active site Lys-131 residue. Here, we have determined the crystal structure of wild-type (WT) λexo in complex with the same DNA substrate, but in the presence of Ca(2+) instead of Mg(2+). Surprisingly, there is only one Ca(2+) bound in the active site, near the position of Mg(A) in the structure with Mg(2+). The scissile phosphate is displaced by 2.2 Å relative to its position in the structure with Mg(2+), and the network of interactions involving the attacking water molecule is broken. Thus, the structure does not represent a catalytic configuration. However, the crystal structure does show clear electron density for the side chain of Lys-131, which is held in place by interactions with Gln-157 and Glu-129. By combining the K131A-Mg(2+) and WT-Ca(2+) structures, we constructed a composite model to show the likely interactions of Lys-131 during catalysis. The implications with regard to the catalytic mechanism are discussed.
- Ohio State Biochemistry Program, ‡Department of Molecular and Cellular Biochemistry, and §Department of Chemistry and Biochemistry, The Ohio State University , 1645 Neil Avenue, Columbus, Ohio 43210, United States.
Organizational Affiliation: