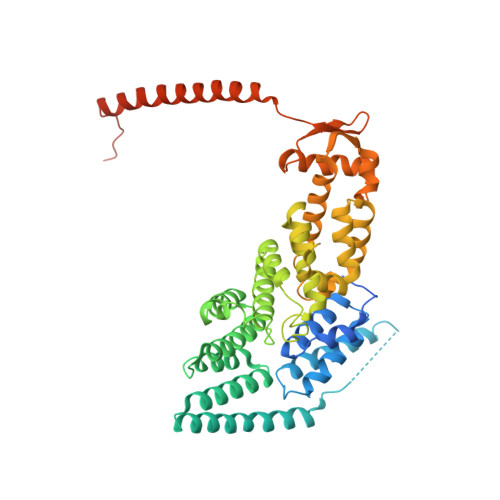
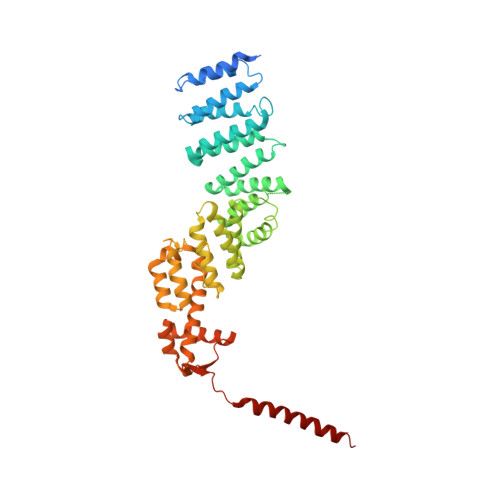
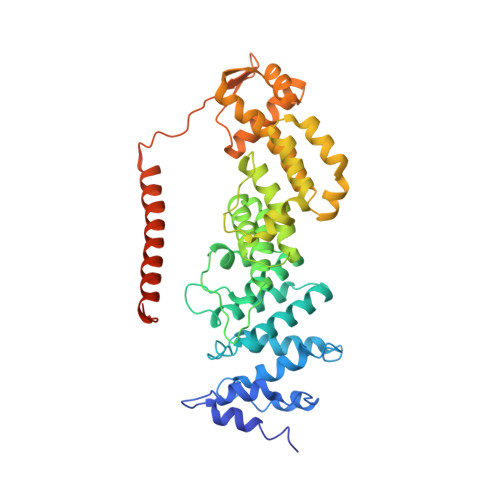
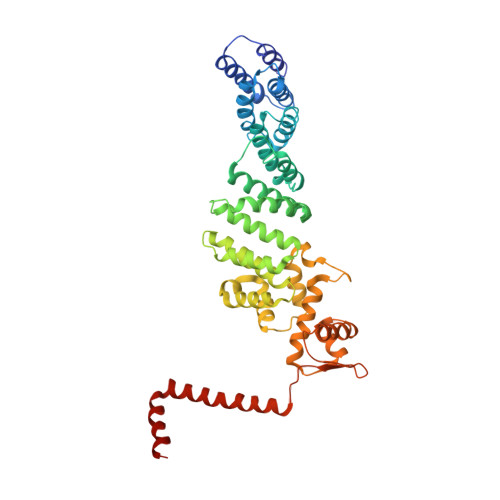
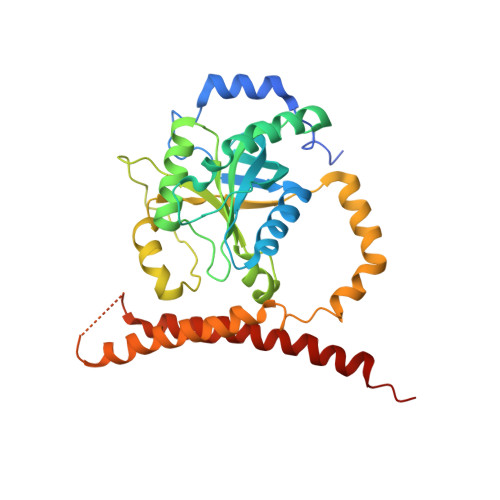
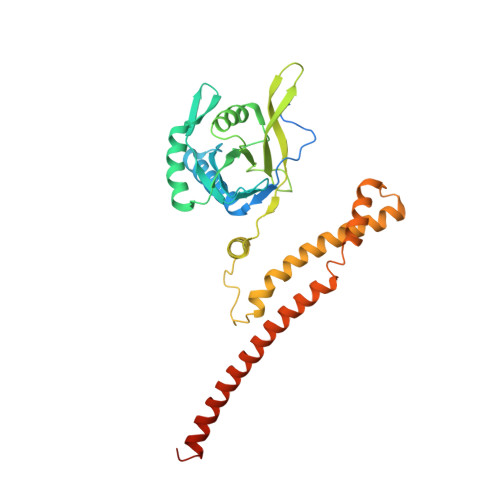
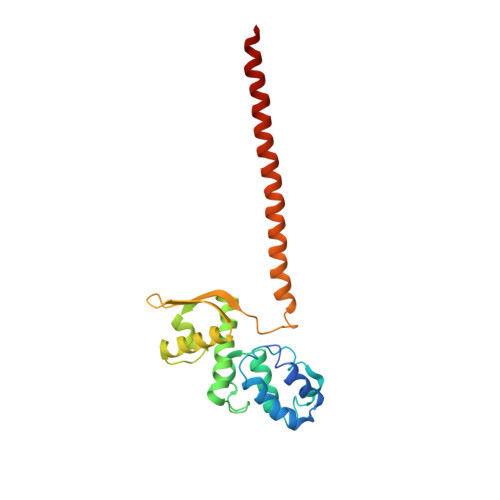
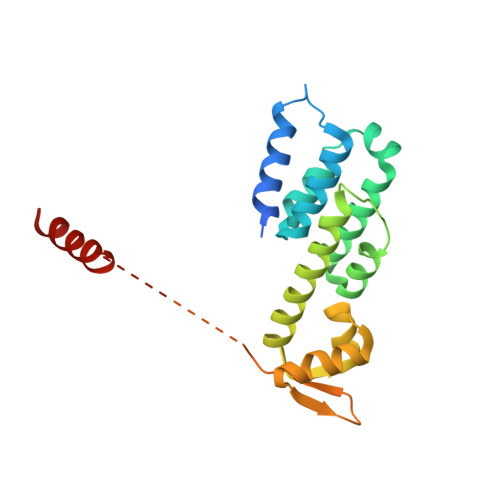
Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome.
Cavadini, S., Fischer, E.S., Bunker, R.D., Potenza, A., Lingaraju, G.M., Goldie, K.N., Mohamed, W.I., Faty, M., Petzold, G., Beckwith, R.E., Tichkule, R.B., Hassiepen, U., Abdulrahman, W., Pantelic, R.S., Matsumoto, S., Sugasawa, K., Stahlberg, H., Thoma, N.H.(2016) Nature 531: 598-603
- PubMed: 27029275
- DOI: https://doi.org/10.1038/nature17416
- Primary Citation of Related Structures:
4WSN - PubMed Abstract:
The cullin-RING ubiquitin E3 ligase (CRL) family comprises over 200 members in humans. The COP9 signalosome complex (CSN) regulates CRLs by removing their ubiquitin-like activator NEDD8. The CUL4A-RBX1-DDB1-DDB2 complex (CRL4A(DDB2)) monitors the genome for ultraviolet-light-induced DNA damage. CRL4A(DBB2) is inactive in the absence of damaged DNA and requires CSN to regulate the repair process. The structural basis of CSN binding to CRL4A(DDB2) and the principles of CSN activation are poorly understood. Here we present cryo-electron microscopy structures for CSN in complex with neddylated CRL4A ligases to 6.4 Å resolution. The CSN conformers defined by cryo-electron microscopy and a novel apo-CSN crystal structure indicate an induced-fit mechanism that drives CSN activation by neddylated CRLs. We find that CSN and a substrate cannot bind simultaneously to CRL4A, favouring a deneddylated, inactive state for substrate-free CRL4 complexes. These architectural and regulatory principles appear conserved across CRL families, allowing global regulation by CSN.
- Friedrich Miescher Institute for Biomedical Research, Maulbeerstrasse 66, 4058 Basel, Switzerland.
Organizational Affiliation: