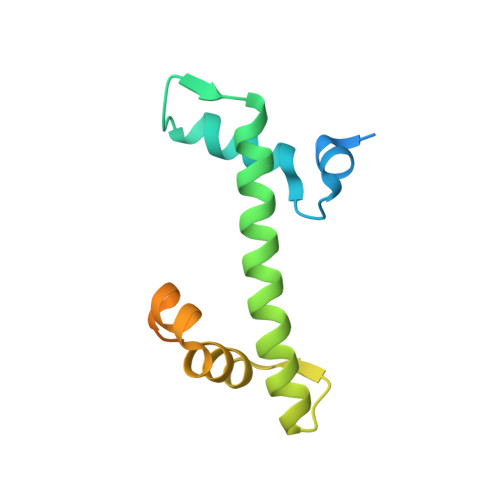
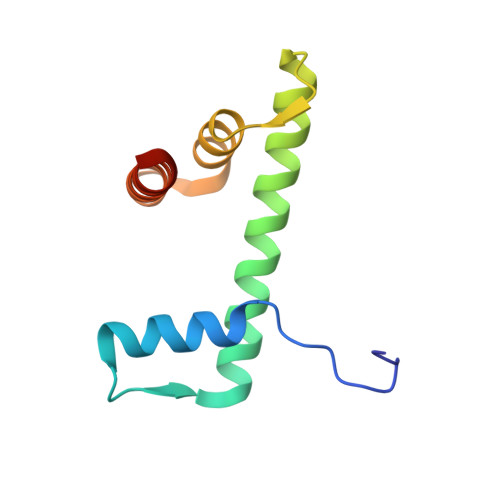
FACT Disrupts Nucleosome Structure by Binding H2A-H2B with Conserved Peptide Motifs.
Kemble, D.J., McCullough, L.L., Whitby, F.G., Formosa, T., Hill, C.P.(2015) Mol Cell 60: 294-306
- PubMed: 26455391
- DOI: https://doi.org/10.1016/j.molcel.2015.09.008
- Primary Citation of Related Structures:
4WNN - PubMed Abstract:
FACT, a heterodimer of Spt16 and Pob3, is an essential histone chaperone. We show that the H2A-H2B binding activity that is central to FACT function resides in short acidic regions near the C termini of each subunit. Mutations throughout these regions affect binding and cause correlated phenotypes that range from mild to lethal, with the largest individual contributions unexpectedly coming from an aromatic residue and a nearby carboxylate residue within each domain. Spt16 and Pob3 bind overlapping sites on H2A-H2B, and Spt16-Pob3 heterodimers simultaneously bind two H2A-H2B dimers, the same stoichiometry as the components of a nucleosome. An Spt16:H2A-H2B crystal structure explains the biochemical and genetic data, provides a model for Pob3 binding, and implies a mechanism for FACT reorganization that we confirm biochemically. Moreover, unexpected similarity to binding of ANP32E and Swr1 with H2A.Z-H2B reveals that diverse H2A-H2B chaperones use common mechanisms of histone binding and regulating nucleosome functions.
- Department of Biochemistry, University of Utah School of Medicine, Salt Lake City, UT 84112-5650, USA.
Organizational Affiliation: