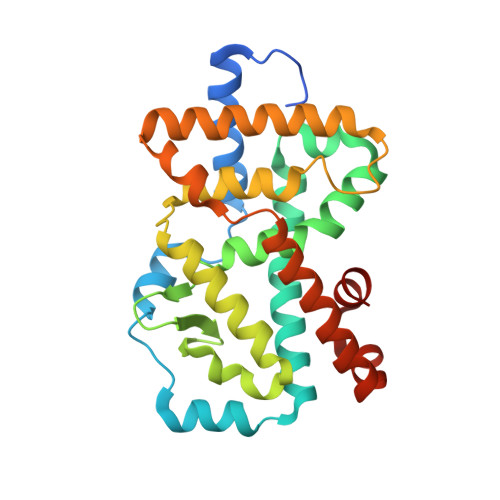
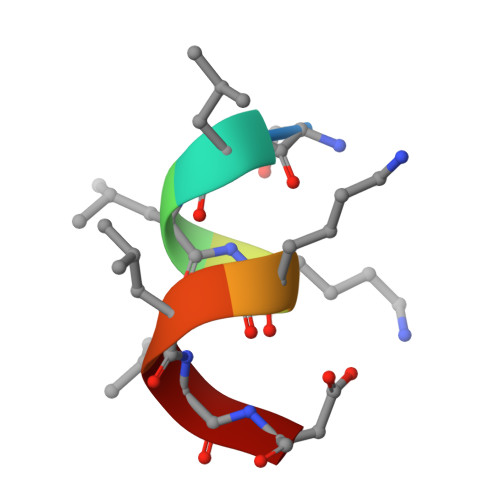
A reversed sulfonamide series of selective RORc inverse agonists.
van Niel, M.B., Fauber, B.P., Cartwright, M., Gaines, S., Killen, J.C., Rene, O., Ward, S.I., de Leon Boenig, G., Deng, Y., Eidenschenk, C., Everett, C., Gancia, E., Ganguli, A., Gobbi, A., Hawkins, J., Johnson, A.R., Kiefer, J.R., La, H., Lockey, P., Norman, M., Ouyang, W., Qin, A., Wakes, N., Waszkowycz, B., Wong, H.(2014) Bioorg Med Chem Lett 24: 5769-5776
- PubMed: 25453817
- DOI: https://doi.org/10.1016/j.bmcl.2014.10.037
- Primary Citation of Related Structures:
4WLB - PubMed Abstract:
The identification of a new series of RORc inverse agonists is described. Comprehensive structure-activity relationship studies of this reversed sulfonamide series identified potent RORc inverse agonists in biochemical and cellular assays which were also selective against a panel of nuclear receptors. Our work has contributed a compound that may serve as a useful in vitro tool to delineate the complex biological pathways involved in signalling through RORc. An X-ray co-crystal structure of an analogue with RORc has also provided useful insights into the binding interactions of the new series.
- Argenta, Early Discovery, Charles River, 7-9 Spire Green Centre, Flex Meadow, Harlow, Essex CM19 5TR, United Kingdom. Electronic address: Bodil.vanniel@crl.com.
Organizational Affiliation: