Altered Peptide Ligands Revisited: Vaccine Design through Chemically Modified HLA-A2-Restricted T Cell Epitopes.
Hoppes, R., Oostvogels, R., Luimstra, J.J., Wals, K., Toebes, M., Bies, L., Ekkebus, R., Rijal, P., Celie, P.H., Huang, J.H., Emmelot, M.E., Spaapen, R.M., Lokhorst, H., Schumacher, T.N., Mutis, T., Rodenko, B., Ovaa, H.(2014) J Immunol 193: 4803-4813
- PubMed: 25311806
- DOI: https://doi.org/10.4049/jimmunol.1400800
- Primary Citation of Related Structures:
4WJ5 - PubMed Abstract:
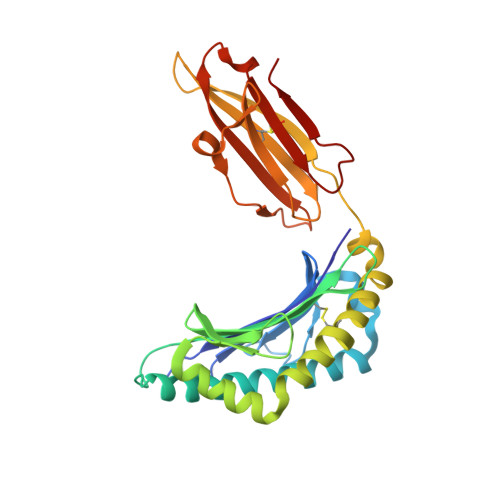
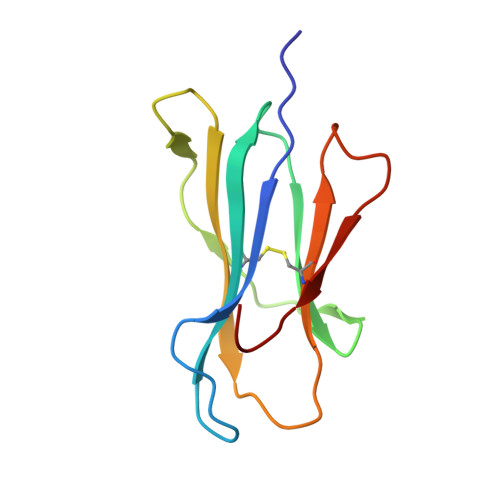
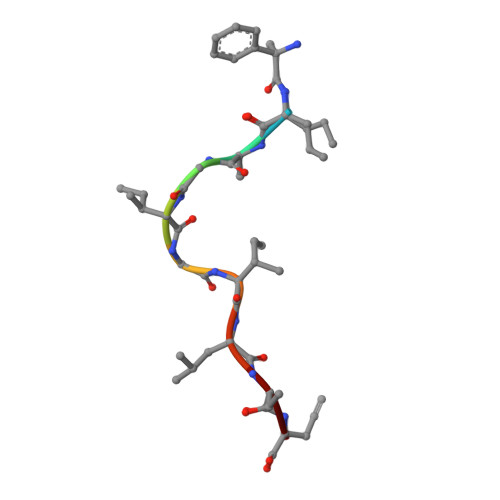
Virus or tumor Ag-derived peptides that are displayed by MHC class I molecules are attractive starting points for vaccine development because they induce strong protective and therapeutic cytotoxic T cell responses. In thus study, we show that the MHC binding and consequent T cell reactivity against several HLA-A*02 restricted epitopes can be further improved through the incorporation of nonproteogenic amino acids at primary and secondary anchor positions. We screened more than 90 nonproteogenic, synthetic amino acids through a range of epitopes and tested more than 3000 chemically enhanced altered peptide ligands (CPLs) for binding affinity to HLA-A*0201. With this approach, we designed CPLs of viral epitopes, of melanoma-associated Ags, and of the minor histocompatibility Ag UTA2-1, which is currently being evaluated for its antileukemic activity in clinical dendritic cell vaccination trials. The crystal structure of one of the CPLs in complex with HLA-A*0201 revealed the molecular interactions likely responsible for improved binding. The best CPLs displayed enhanced affinity for MHC, increasing MHC stability and prolonging recognition by Ag-specific T cells and, most importantly, they induced accelerated expansion of antitumor T cell frequencies in vitro and in vivo as compared with the native epitope. Eventually, we were able to construct a toolbox of preferred nonproteogenic residues with which practically any given HLA-A*02 restricted epitope can be readily optimized. These CPLs could improve the therapeutic outcome of vaccination strategies or can be used for ex vivo enrichment and faster expansion of Ag-specific T cells for transfer into patients.
- Division of Cell Biology, The Netherlands Cancer Institute, 1066 CX Amsterdam, the Netherlands;
Organizational Affiliation: