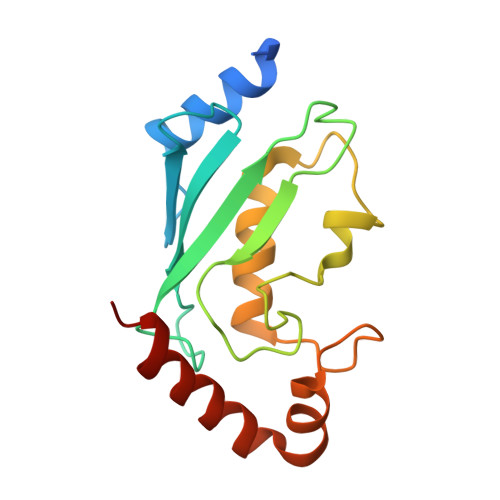
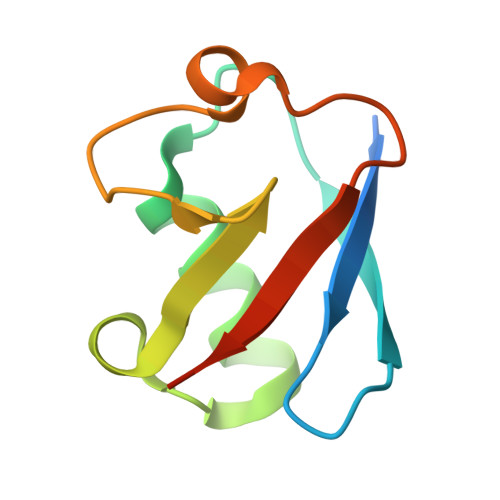
RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment.
Hodge, C.D., Ismail, I.H., Edwards, R.A., Hura, G.L., Xiao, A.T., Tainer, J.A., Hendzel, M.J., Glover, J.N.(2016) J Biological Chem 291: 9396-9410
- PubMed: 26903517
- DOI: https://doi.org/10.1074/jbc.M116.715698
- Primary Citation of Related Structures:
4WHV - PubMed Abstract:
DNA double strand break (DSB) responses depend on the sequential actions of the E3 ubiquitin ligases RNF8 and RNF168 plus E2 ubiquitin-conjugating enzyme Ubc13 to specifically generate histone Lys-63-linked ubiquitin chains in DSB signaling. Here, we defined the activated RNF8-Ubc13∼ubiquitin complex by x-ray crystallography and its functional solution conformations by x-ray scattering, as tested by separation-of-function mutations imaged in cells by immunofluorescence. The collective results show that the RING E3 RNF8 targets E2 Ubc13 to DSB sites and plays a critical role in damage signaling by stimulating polyubiquitination through modulating conformations of ubiquitin covalently linked to the Ubc13 active site. Structure-guided separation-of-function mutations show that the RNF8 E2 stimulating activity is essential for DSB signaling in mammalian cells and is necessary for downstream recruitment of 53BP1 and BRCA1. Chromatin-targeted RNF168 rescues 53BP1 recruitment involved in non-homologous end joining but not BRCA1 recruitment for homologous recombination. These findings suggest an allosteric approach to targeting the ubiquitin-docking cleft at the E2-E3 interface for possible interventions in cancer and chronic inflammation, and moreover, they establish an independent RNF8 role in BRCA1 recruitment.
- From the Department of Biochemistry, University of Alberta, Edmonton, Alberta T6G 2H7, Canada.
Organizational Affiliation: