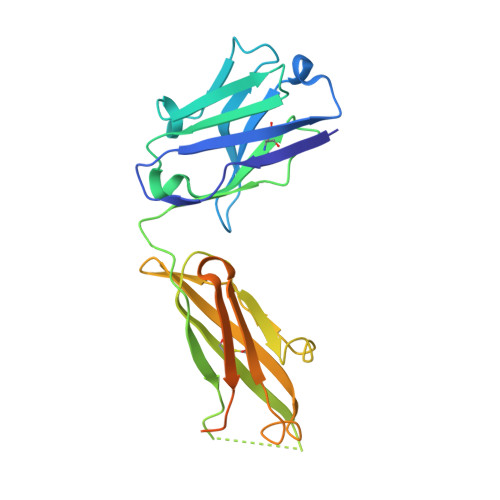
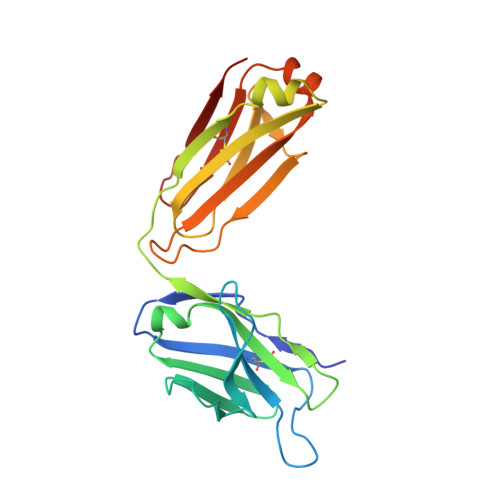
Structural flexibility of a conserved antigenic region in hepatitis C virus glycoprotein e2 recognized by broadly neutralizing antibodies.
Meola, A., Tarr, A.W., England, P., Meredith, L.W., McClure, C.P., Foung, S.K., McKeating, J.A., Ball, J.K., Rey, F.A., Krey, T.(2015) J Virol 89: 2170-2181
- PubMed: 25473061
- DOI: https://doi.org/10.1128/JVI.02190-14
- Primary Citation of Related Structures:
4WHT, 4WHY - PubMed Abstract:
Neutralizing antibodies (NAbs) targeting glycoprotein E2 are important for the control of hepatitis C virus (HCV) infection. One conserved antigenic site (amino acids 412 to 423) is disordered in the reported E2 structure, but a synthetic peptide mimicking this site forms a β-hairpin in complex with three independent NAbs. Our structure of the same peptide in complex with NAb 3/11 demonstrates a strikingly different extended conformation. We also show that residues 412 to 423 are essential for virus entry but not for E2 folding. Together with the neutralizing capacity of the 3/11 Fab fragment, this indicates an unexpected structural flexibility within this epitope. NAbs 3/11 and AP33 (recognizing the extended and β-hairpin conformations, respectively) display similar neutralizing activities despite converse binding kinetics. Our results suggest that HCV utilizes conformational flexibility as an immune evasion strategy, contributing to the limited immunogenicity of this epitope in patients, similar to the conformational flexibility described for other enveloped and nonenveloped viruses. Approximately 180 million people worldwide are infected with hepatitis C virus (HCV), and neutralizing antibodies play an important role in controlling the replication of this major human pathogen. We show here that one of the most conserved antigenic sites within the major glycoprotein E2 (amino acids 412 to 423), which is disordered in the recently reported crystal structure of an E2 core fragment, can adopt different conformations in the context of the infectious virus particle. Recombinant Fab fragments recognizing different conformations of this antigenic site have similar neutralization activities in spite of converse kinetic binding parameters. Of note, an antibody response targeting this antigenic region is less frequent than those targeting other more immunogenic regions in E2. Our results suggest that the observed conformational flexibility in this conserved antigenic region contributes to the evasion of the humoral host immune response, facilitating chronicity and the viral spread of HCV within an infected individual.
- Institut Pasteur, Unité de Virologie Structurale, Department Virologie, Paris, France CNRS UMR 3569, Paris, France.
Organizational Affiliation: