X-ray Crystallographic Structure of BshC, a Unique Enzyme Involved in Bacillithiol Biosynthesis.
VanDuinen, A.J., Winchell, K.R., Keithly, M.E., Cook, P.D.(2015) Biochemistry 54: 100-103
- PubMed: 25496067
- DOI: https://doi.org/10.1021/bi501394q
- Primary Citation of Related Structures:
4WBD - PubMed Abstract:
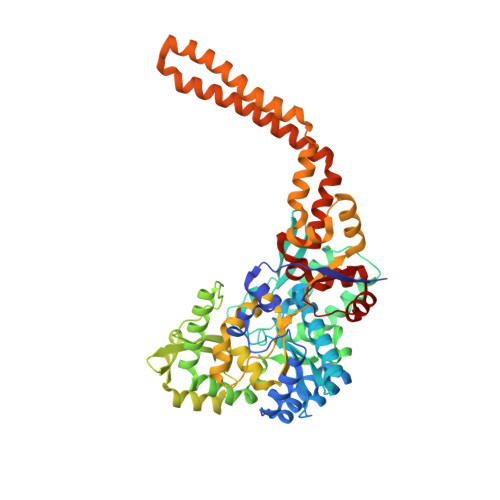
Bacillithiol is produced by many Gram-positive bacteria via a pathway utilizing the enzymes BshA, BshB, and BshC. Here we report the 1.77 Å resolution crystal structure of BshC, the putative cysteine ligase in bacillithiol production. The structure reveals that BshC contains a core Rossmann fold with connecting peptide motifs (CP1 and CP2) and a unique α-helical coiled-coil domain that facilitates dimerization. The model contains citrate and glycerol in the canonical active site and ADP in a second binding pocket. The overall structure and bound ligands give insight into the function of this unique enzyme.
- Department of Cell & Molecular Biology, Grand Valley State University , Allendale, Michigan 49401, United States.
Organizational Affiliation: