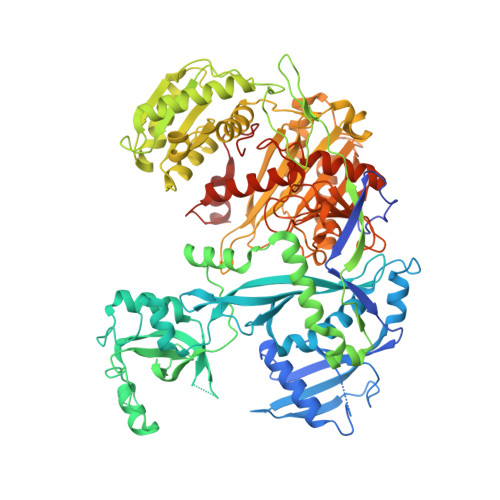
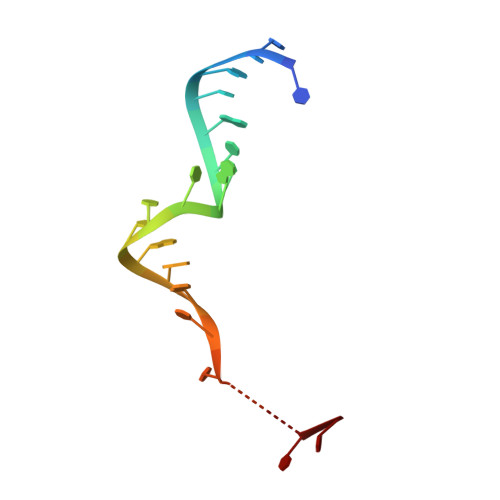
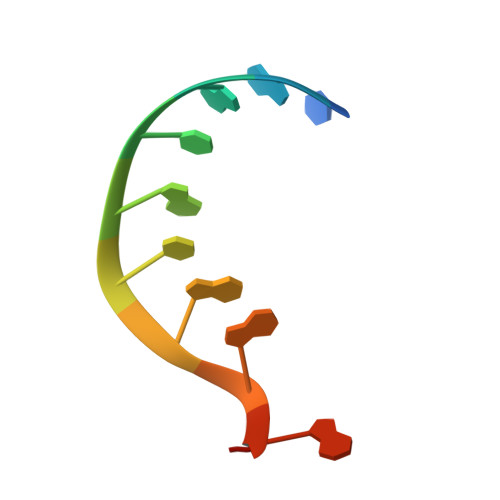
Gene regulation. Structural basis for microRNA targeting.
Schirle, N.T., Sheu-Gruttadauria, J., MacRae, I.J.(2014) Science 346: 608-613
- PubMed: 25359968
- DOI: https://doi.org/10.1126/science.1258040
- Primary Citation of Related Structures:
4W5N, 4W5O, 4W5Q, 4W5R, 4W5T - PubMed Abstract:
MicroRNAs (miRNAs) control expression of thousands of genes in plants and animals. miRNAs function by guiding Argonaute proteins to complementary sites in messenger RNAs (mRNAs) targeted for repression. We determined crystal structures of human Argonaute-2 (Ago2) bound to a defined guide RNA with and without target RNAs representing miRNA recognition sites. These structures suggest a stepwise mechanism, in which Ago2 primarily exposes guide nucleotides (nt) 2 to 5 for initial target pairing. Pairing to nt 2 to 5 promotes conformational changes that expose nt 2 to 8 and 13 to 16 for further target recognition. Interactions with the guide-target minor groove allow Ago2 to interrogate target RNAs in a sequence-independent manner, whereas an adenosine binding-pocket opposite guide nt 1 further facilitates target recognition. Spurious slicing of miRNA targets is avoided through an inhibitory coordination of one catalytic magnesium ion. These results explain the conserved nucleotide-pairing patterns in animal miRNA target sites first observed over two decades ago.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: