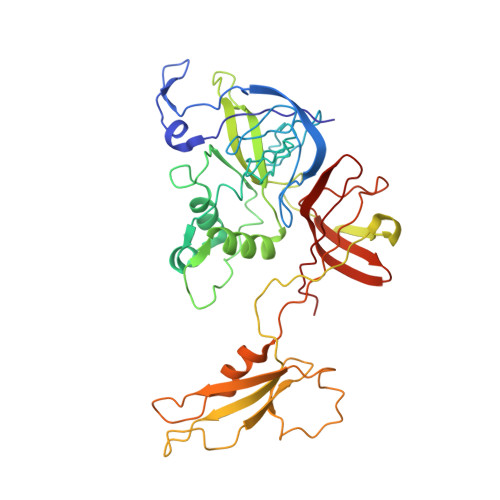
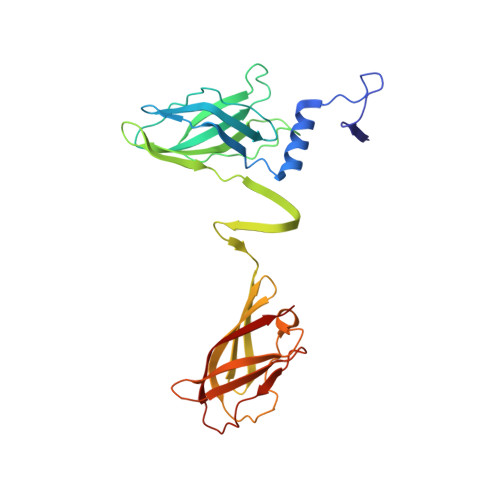
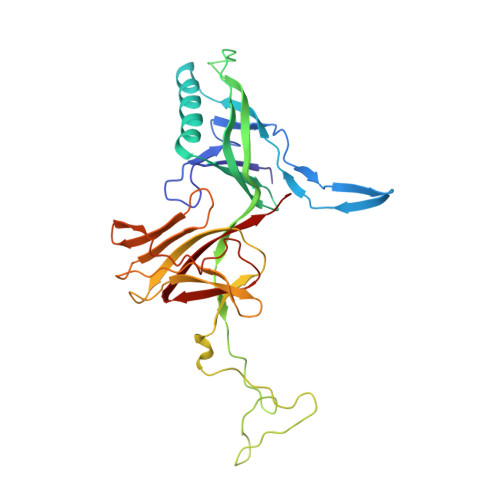
Structure of Lactococcal Phage P2 Baseplate and its Mechanism of Activation.
Sciara, G., Bebeacua, C., Bron, P., Tremblay, D., Ortiz-Lombardia, M., Lichiere, J., van Heel, M., Campanacci, V., Moineau, S., Cambillau, C.(2010) Proc Natl Acad Sci U S A 107: 6852
- PubMed: 20351260
- DOI: https://doi.org/10.1073/pnas.1000232107
- Primary Citation of Related Structures:
2WZP, 2X53, 4V5I - PubMed Abstract:
Siphoviridae is the most abundant viral family on earth which infects bacteria as well as archaea. All known siphophages infecting gram+ Lactococcus lactis possess a baseplate at the tip of their tail involved in host recognition and attachment. Here, we report analysis of the p2 phage baseplate structure by X-ray crystallography and electron microscopy and propose a mechanism for the baseplate activation during attachment to the host cell. This approximately 1 MDa, Escherichia coli-expressed baseplate is composed of three protein species, including six trimers of the receptor-binding protein (RBP). RBPs host-recognition domains point upwards, towards the capsid, in agreement with the electron-microscopy map of the free virion. In the presence of Ca(2+), a cation mandatory for infection, the RBPs rotated 200 degrees downwards, presenting their binding sites to the host, and a channel opens at the bottom of the baseplate for DNA passage. These conformational changes reveal a novel siphophage activation and host-recognition mechanism leading ultimately to DNA ejection.
- Architecture et Fonction des Macromolécules Biologiques, UMR 6098 Centre National de la Recherche Scientifique and Universités d'Aix-Marseille I & II, Campus de Luminy, Case 932, 13288 Marseille Cedex 09, France.
Organizational Affiliation: