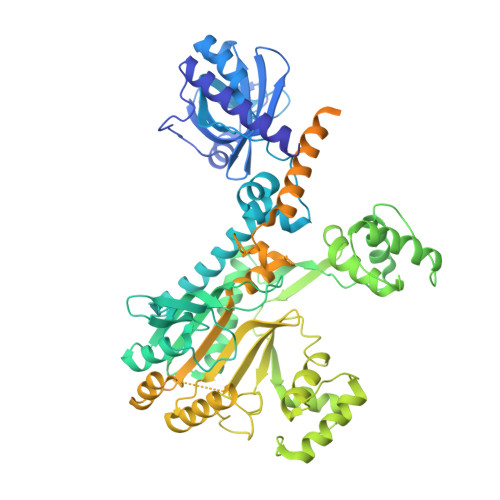
Crystal Structures of Entamoeba Histolytica Lysyl-tRNA Synthetase Reveal Conformational Changes Upon Lysine Binding and a Specific Helix Bundle Domain.
Bonnefond, L., Castro De Moura, M., Ribas De Pouplana, L., Nureki, O.(2014) FEBS Lett 588: 4478
- PubMed: 25448989
- DOI: https://doi.org/10.1016/j.febslet.2014.10.019
- Primary Citation of Related Structures:
4UP7, 4UP8, 4UP9, 4UPA - PubMed Abstract:
The class II lysyl-tRNA synthetases (KRS) are conserved aminoacyl-tRNA synthetases that attach lysine to the cognate tRNA in a two-step mechanism. The enzyme from the parasitic protozoan Entamoeba histolytica was crystallized in the presence of small ligands to generate snapshots of the lysine-adenylate formation. The residues involved in lysine activation are highly conserved and the active site closes around the lysyl-adenylate, as observed in bacterial KRS. The Entamoeba EMAPII-like polypeptide is not resolved in the crystals, but another Entamoeba-specific insertion could be modeled as a small helix bundle that may contribute to tRNA binding through interaction with the tRNA hinge.
- Department of Biophysics and Biochemistry, Graduate School of Science, The University of Tokyo, 2-11-16, Yayoi, Bunkyo, Tokyo 113-0032, Japan. lbonnefond@unistra.fr
Organizational Affiliation: