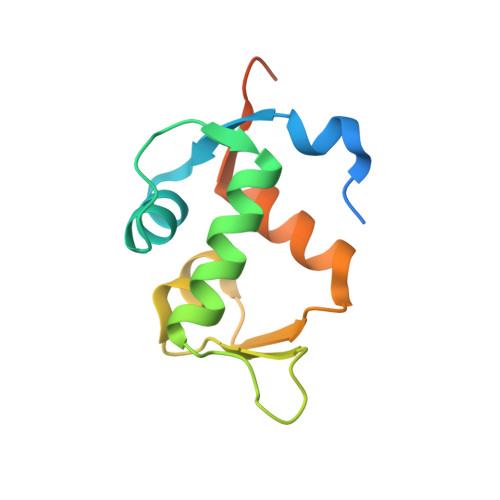
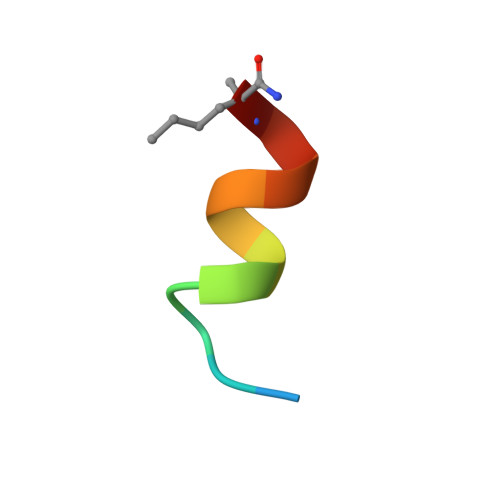
Structure of a stapled peptide antagonist bound to nutlin-resistant Mdm2.
Chee, S.M., Wongsantichon, J., Soo Tng, Q., Robinson, R., Joseph, T.L., Verma, C., Lane, D.P., Brown, C.J., Ghadessy, F.J.(2014) PLoS One 9: e104914-e104914
- PubMed: 25115702
- DOI: https://doi.org/10.1371/journal.pone.0104914
- Primary Citation of Related Structures:
4UMN - PubMed Abstract:
As key negative regulator of the p53 tumour suppressor, Mdm2 is an attractive therapeutic target. Small molecules such as Nutlin have been developed to antagonise Mdm2, resulting in p53-dependent death of tumour cells. We have recently described a mutation in Mdm2 (M62A), which precludes binding of Nutlin, but not p53. This Nutlin-resistant variant is not, however, refractory to binding and inhibition by stapled peptide antagonists targeting the same region of Mdm2. A detailed understanding of how stapled peptides are recalcitrant to Mdm2 mutations conferring Nutlin-resistance will aid in the further development of potent Mdm2 antagonists. Here, we report the 2.00 Å crystal structure of a stapled peptide antagonist bound to Nutlin resistant Mdm2. The stapled peptide relies on an extended network of interactions along the hydrophobic binding cleft of Mdm2 for high affinity binding. Additionally, as seen in other stapled peptide structures, the hydrocarbon staple itself contributes to binding through favourable interactions with Mdm2. The structure highlights the intrinsic plasticity present in both Mdm2 and the hydrocarbon staple moiety, and can be used to guide future iterations of both small molecules and stapled peptides for improved antagonists of Mdm2.
- p53Lab, Agency for Science Technology and Research (A*STAR), Singapore, Singapore.
Organizational Affiliation: