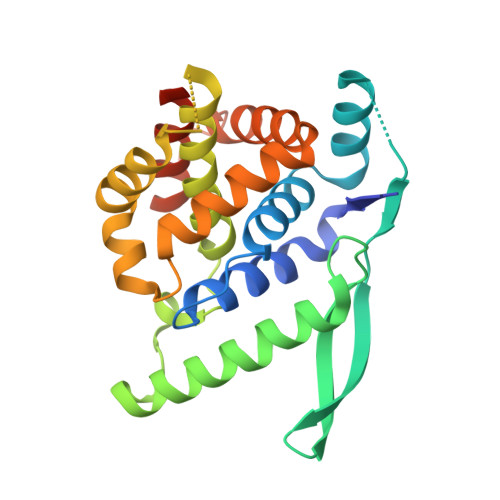
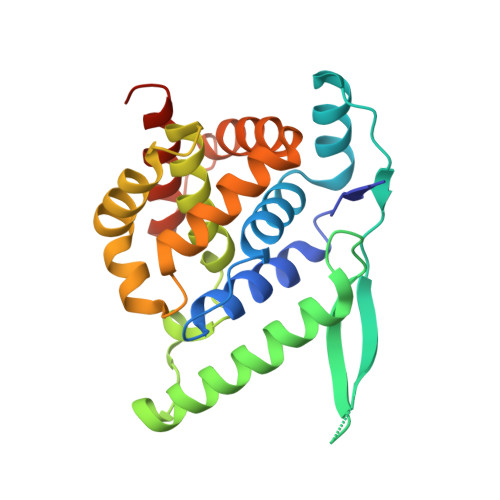
A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
Ouizougun-Oubari, M., Pereira, N., Tarus, B., Galloux, M., Lassoued, S., Fix, J., Tortorici, M.A., Hoos, S., Baron, B., England, P., Desmaele, D., Couvreur, P., Bontems, F., Rey, F.A., Eleouet, J.F., Sizun, C., Slama-Schwok, A., Duquerroy, S.(2015) J Virol 89: 11129
- PubMed: 26246564
- DOI: https://doi.org/10.1128/JVI.01612-15
- Primary Citation of Related Structures:
4UC6, 4UC7, 4UC8, 4UC9, 4UCA, 4UCB, 4UCC, 4UCD, 4UCE - PubMed Abstract:
Presently, respiratory syncytial virus (RSV), the main cause of severe respiratory infections in infants, cannot be treated efficiently with antivirals. However, its RNA-dependent polymerase complex offers potential targets for RSV-specific drugs. This includes the recognition of its template, the ribonucleoprotein complex (RNP), consisting of genomic RNA encapsidated by the RSV nucleoprotein, N. This recognition proceeds via interaction between the phosphoprotein P, which is the main polymerase cofactor, and N. The determinant role of the C terminus of P, and more particularly of the last residue, F241, in RNP binding and viral RNA synthesis has been assessed previously. Here, we provide detailed structural insight into this crucial interaction for RSV polymerase activity. We solved the crystallographic structures of complexes between the N-terminal domain of N (N-NTD) and C-terminal peptides of P and characterized binding by biophysical approaches. Our results provide a rationale for the pivotal role of F241, which inserts into a well-defined N-NTD pocket. This primary binding site is completed by transient contacts with upstream P residues outside the pocket. Based on the structural information of the N-NTD:P complex, we identified inhibitors of this interaction, selected by in silico screening of small compounds, that efficiently bind to N and compete with P in vitro. One of the compounds displayed inhibitory activity on RSV replication, thereby strengthening the relevance of N-NTD for structure-based design of RSV-specific antivirals. Respiratory syncytial virus (RSV) is a widespread pathogen that is a leading cause of acute lower respiratory infections in infants worldwide. RSV cannot be treated efficiently with antivirals, and no vaccine is presently available, with the development of pediatric vaccines being particularly challenging. Therefore, there is a need for new therapeutic strategies that specifically target RSV. The interaction between the RSV phosphoprotein P and the ribonucleoprotein complex is critical for viral replication. In this study, we identified the main structural determinants of this interaction, and we used them to screen potential inhibitors in silico. We found a family of molecules that were efficient competitors of P in vitro and showed inhibitory activity on RSV replication in cellular assays. These compounds provide a basis for a pharmacophore model that must be improved but that holds promises for the design of new RSV-specific antivirals.
- Institut Pasteur, Département de Virologie, Unité de Virologie Structurale, Paris, France CNRS UMR 3569 Virologie, Paris, France.
Organizational Affiliation: