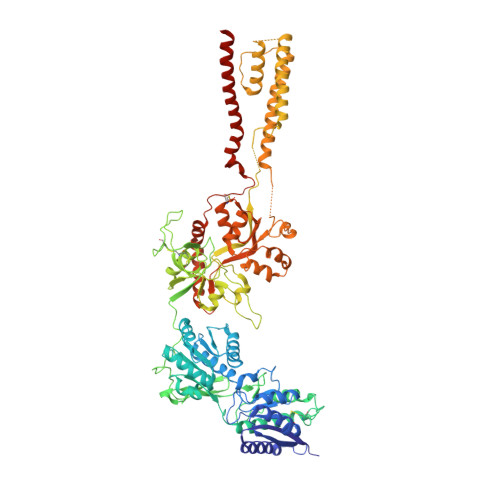
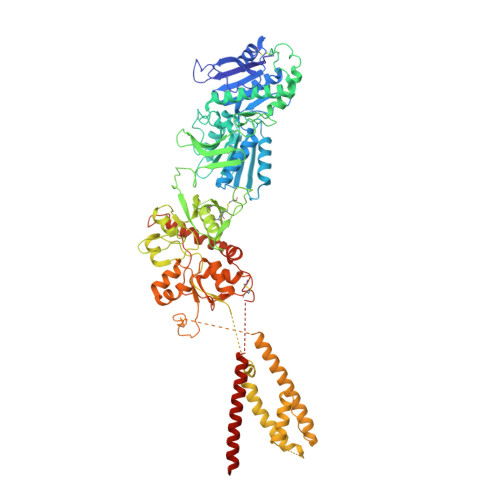
NMDA receptor structures reveal subunit arrangement and pore architecture.
Lee, C.H., Lu, W., Michel, J.C., Goehring, A., Du, J., Song, X., Gouaux, E.(2014) Nature 511: 191-197
- PubMed: 25008524
- DOI: https://doi.org/10.1038/nature13548
- Primary Citation of Related Structures:
4TLL, 4TLM - PubMed Abstract:
N-methyl-d-aspartate (NMDA) receptors are Hebbian-like coincidence detectors, requiring binding of glycine and glutamate in combination with the relief of voltage-dependent magnesium block to open an ion conductive pore across the membrane bilayer. Despite the importance of the NMDA receptor in the development and function of the brain, a molecular structure of an intact receptor has remained elusive. Here we present X-ray crystal structures of the Xenopus laevis GluN1-GluN2B NMDA receptor with the allosteric inhibitor, Ro25-6981, partial agonists and the ion channel blocker, MK-801. Receptor subunits are arranged in a 1-2-1-2 fashion, demonstrating extensive interactions between the amino-terminal and ligand-binding domains. The transmembrane domains harbour a closed-blocked ion channel, a pyramidal central vestibule lined by residues implicated in binding ion channel blockers and magnesium, and a ∼twofold symmetric arrangement of ion channel pore loops. These structures provide new insights into the architecture, allosteric coupling and ion channel function of NMDA receptors.
- 1] Vollum Institute, Oregon Health & Science University, 3181 SW Sam Jackson Park Road, Portland, Oregon 97239, USA [2].
Organizational Affiliation: