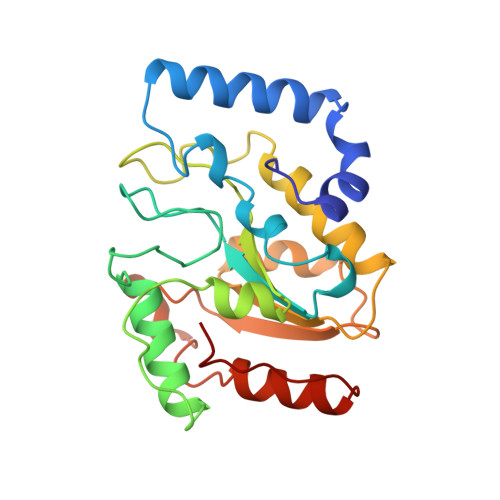
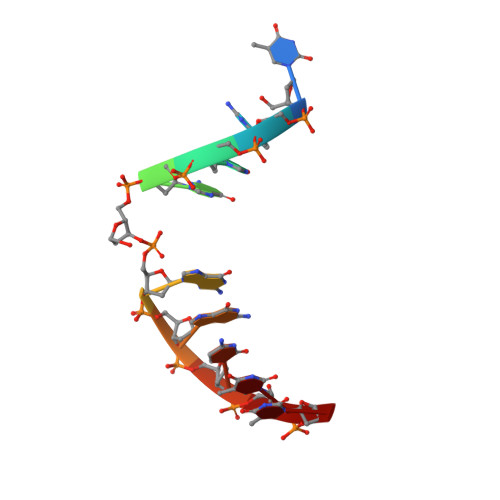
A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA.
Slupphaug, G., Mol, C.D., Kavli, B., Arvai, A.S., Krokan, H.E., Tainer, J.A.(1996) Nature 384: 87-92
- PubMed: 8900285
- DOI: https://doi.org/10.1038/384087a0
- Primary Citation of Related Structures:
4SKN - PubMed Abstract:
Any uracil bases in DNA, a result of either misincorporation or deamination of cytosine, are removed by uracil-DNA glycosylase (UDG), one of the most efficient and specific of the base-excision DNA-repair enzymes. Crystal structures of human and viral UDGs complexed with free uracil have indicated that the enzyme binds an extrahelical uracil. Such binding of undamaged extrahelical bases has been seen in the structures of two bacterial methyltransferases and bacteriophage T4 endonuclease V. Here we characterize the DNA binding and kinetics of several engineered human UDG mutants and present the crystal structure of one of these, which to our knowledge represents the first structure of any eukaryotic DNA repair enzyme in complex with its damaged, target DNA. Electrostatic orientation along the UDG active site, insertion of an amino acid (residue 272) into the DNA through the minor groove, and compression of the DNA backbone flanking the uracil all result in the flipping-out of the damaged base from the DNA major groove, allowing specific recognition of its phosphate, deoxyribose and uracil moieties. Our structure thus provides a view of a productive complex specific for cleavage of uracil from DNA and also reveals the basis for the enzyme-assisted nucleotide flipping by this critical DNA-repair enzyme.
- UNIGEN Center for Molecular Biology, The Norwegian University of Science and Technology, Trondheim, Norway.
Organizational Affiliation: