Effects of the regions adjacent to the RGD motif in disintegrins on their inhibitory activities and structures
Huang, C.H., Shiu, J.H., Chang, Y.T., Jeng, W.Y., Chuang, W.J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Disintegrin rhodostomin | 68 | Calloselasma rhodostoma | Mutation(s): 1 Gene Names: RHOD | 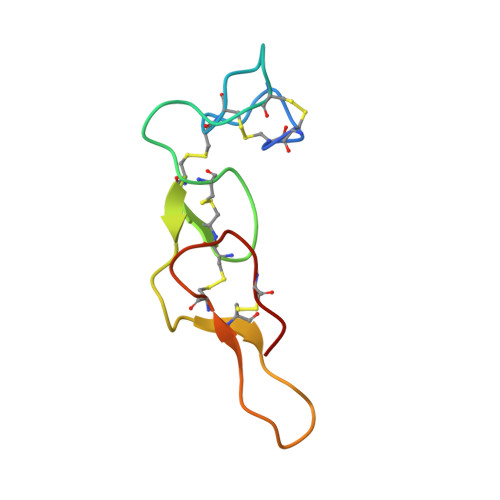 | |
UniProt | |||||
Find proteins for P30403 (Calloselasma rhodostoma) Explore P30403 Go to UniProtKB: P30403 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P30403 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 29.01 | α = 90 |
| b = 74.046 | β = 115.55 |
| c = 29.01 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data collection |
| PHASER | phasing |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |