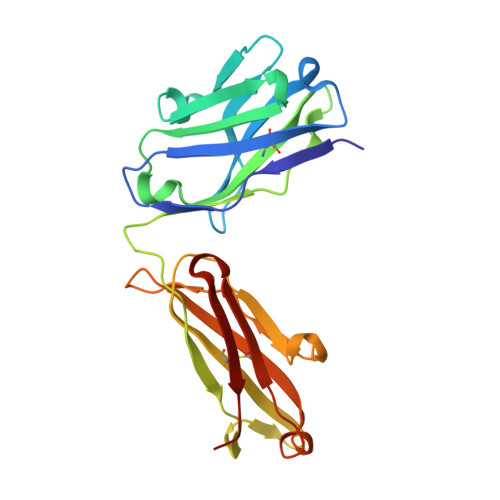
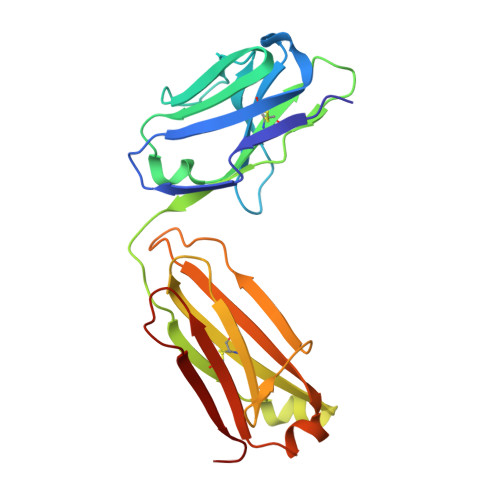
Molecular basis for antagonistic activity of anifrolumab, an anti-interferon-alpha receptor 1 antibody.
Peng, L., Oganesyan, V., Wu, H., Dall'Acqua, W.F., Damschroder, M.M.(2015) MAbs 7: 428-439
- PubMed: 25606664
- DOI: https://doi.org/10.1080/19420862.2015.1007810
- Primary Citation of Related Structures:
4QXG - PubMed Abstract:
Anifrolumab (anifrolumab) is an antagonist human monoclonal antibody that targets interferon α receptor 1 (IFNAR1). Anifrolumab has been developed to treat autoimmune diseases and is currently in clinical trials. To decipher the molecular basis of its mechanism of action, we engaged in multiple epitope mapping approaches to determine how it interacts with IFNAR1 and antagonizes the receptor. We identified the epitope of anifrolumab using enzymatic fragmentation, phage-peptide library panning and mutagenesis approaches. Our studies revealed that anifrolumab recognizes the SD3 subdomain of IFNAR1 with the critical residue R(279). Further, we solved the crystal structure of anifrolumab Fab to a resolution of 2.3 Å. Guided by our epitope mapping studies, we then used in silico protein docking of the anifrolumab Fab crystal structure to IFNAR1 and characterized the corresponding mode of binding. We find that anifrolumab sterically inhibits the binding of IFN ligands to IFNAR1, thus blocking the formation of the ternary IFN/IFNAR1/IFNAR2 signaling complex. This report provides the molecular basis for the mechanism of action of anifrolumab and may provide insights toward designing antibody therapies against IFNAR1.
- a Department of Antibody Discovery and Protein Engineering ; MedImmune LLC ; Gaithersburg , MD USA.
Organizational Affiliation: