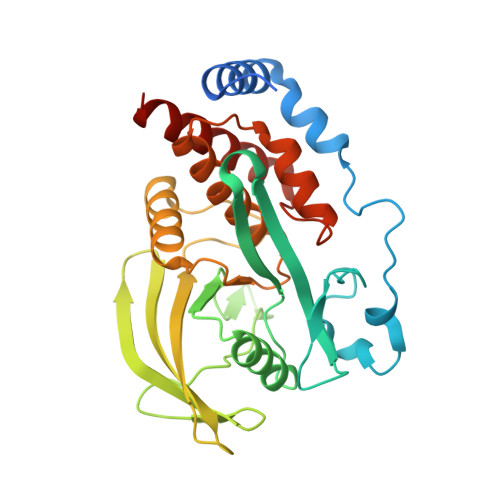
Reciprocal allosteric regulation of p38 gamma and PTPN3 involves a PDZ domain-modulated complex formation.
Chen, K.E., Lin, S.Y., Wu, M.J., Ho, M.R., Santhanam, A., Chou, C.C., Meng, T.C., Wang, A.H.J.(2014) Sci Signal 7: ra98-ra98
- PubMed: 25314968
- DOI: https://doi.org/10.1126/scisignal.2005722
- Primary Citation of Related Structures:
4QUM, 4QUN - PubMed Abstract:
The mitogen-activated protein kinase p38γ (also known as MAPK12) and its specific phosphatase PTPN3 (also known as PTPH1) cooperate to promote Ras-induced oncogenesis. We determined the architecture of the PTPN3-p38γ complex by a hybrid method combining x-ray crystallography, small-angle x-ray scattering, and chemical cross-linking coupled to mass spectrometry. A unique feature of the glutamic acid-containing loop (E-loop) of the phosphatase domain defined the substrate specificity of PTPN3 toward fully activated p38γ. The solution structure revealed the formation of an active-state complex between p38γ and the phosphatase domain of PTPN3. The PDZ domain of PTPN3 stabilized the active-state complex through an interaction with the PDZ-binding motif of p38γ. This interaction alleviated autoinhibition of PTPN3, enabling efficient tyrosine dephosphorylation of p38γ. Our findings may enable structure-based drug design targeting the PTPN3-p38γ interaction as an anticancer therapeutic.
- Institute of Biological Chemistry, Academia Sinica, Taipei 11581, Taiwan.
Organizational Affiliation: