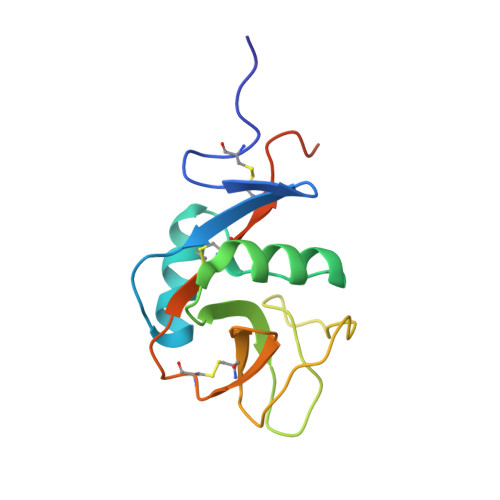
Four crystal structures of human LLT1, a ligand of human NKR-P1, in varied glycosylation and oligomerization states
Skalova, T., Blaha, J., Harlos, K., Duskova, J., Koval, T., Stransky, J., Hasek, J., Vanek, O., Dohnalek, J.(2015) Acta Crystallogr D Biol Crystallogr 71: 578-591
- PubMed: 25760607
- DOI: https://doi.org/10.1107/S1399004714027928
- Primary Citation of Related Structures:
4QKG, 4QKH, 4QKI, 4QKJ - PubMed Abstract:
Human LLT1 is a C-type lectin-like ligand of NKR-P1 (CD161, gene KLRB1), a C-type lectin-like receptor of natural killer cells. Using X-ray diffraction, the first experimental structures of human LLT1 were determined. Four structures of LLT1 under various conditions were determined: monomeric, dimeric deglycosylated after the first N-acetylglucosamine unit in two forms and hexameric with homogeneous GlcNAc2Man5 glycosylation. The dimeric form follows the classical dimerization mode of human CD69. The monomeric form keeps the same fold with the exception of the position of an outer part of the long loop region. The hexamer of glycosylated LLT1 consists of three classical dimers. The hexameric packing may indicate a possible mode of interaction of C-type lectin-like proteins in the glycosylated form.
- Institute of Biotechnology, Academy of Sciences of the Czech Republic, v.v.i., Vídeňská 1083, 142 20 Praha 4, Czech Republic.
Organizational Affiliation: