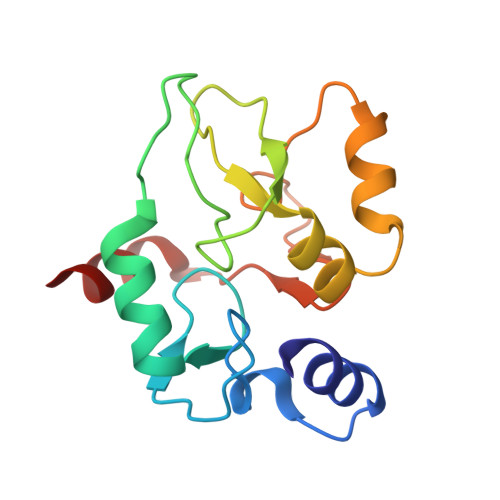
Engineering of a histone-recognition domain in a de novo DNA methyltransferase alters the epigenetic landscape of ESCs
Noh, K., Wang, H., Kim, H., Wenderski, W., Fang, F., Li, C., Dewell, S., Wu, X., Ferris, A., Hughes, S.H., Zheng, D., Melnick, A.M., Patel, D.J., Li, H., Allis, C.D.To be published.