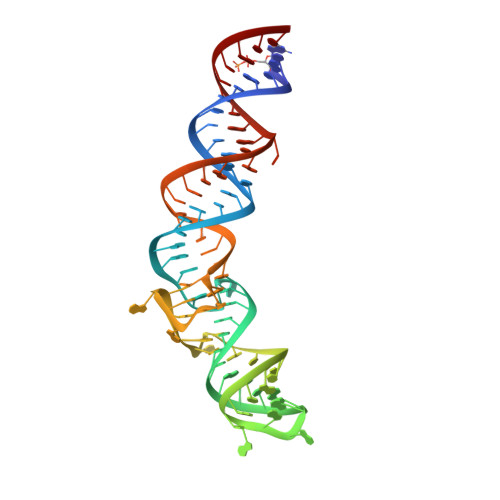
A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Huang, H., Suslov, N.B., Li, N.S., Shelke, S.A., Evans, M.E., Koldobskaya, Y., Rice, P.A., Piccirilli, J.A.(2014) Nat Chem Biol 10: 686-691
- PubMed: 24952597
- DOI: https://doi.org/10.1038/nchembio.1561
- Primary Citation of Related Structures:
4KZD, 4KZE, 4Q9Q, 4Q9R - PubMed Abstract:
Spinach is an in vitro-selected RNA aptamer that binds a GFP-like ligand and activates its green fluorescence. Spinach is thus an RNA analog of GFP and has potentially widespread applications for in vivo labeling and imaging. We used antibody-assisted crystallography to determine the structures of Spinach both with and without bound fluorophore at 2.2-Å and 2.4-Å resolution, respectively. Spinach RNA has an elongated structure containing two helical domains separated by an internal bulge that folds into a G-quadruplex motif of unusual topology. The G-quadruplex motif and adjacent nucleotides comprise a partially preformed binding site for the fluorophore. The fluorophore binds in a planar conformation and makes extensive aromatic stacking and hydrogen bond interactions with the RNA. Our findings provide a foundation for structure-based engineering of new fluorophore-binding RNA aptamers.
- Department of Chemistry, University of Chicago, Chicago, Illinois, USA.
Organizational Affiliation: