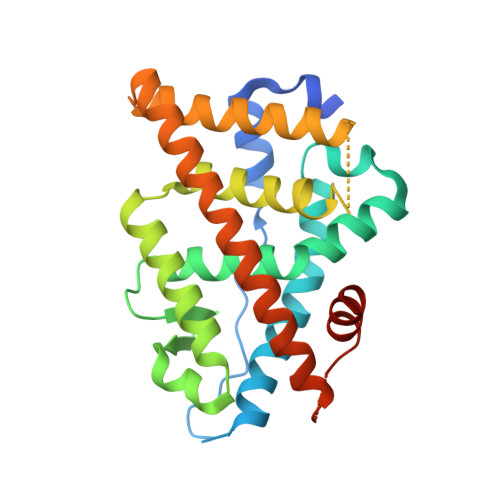
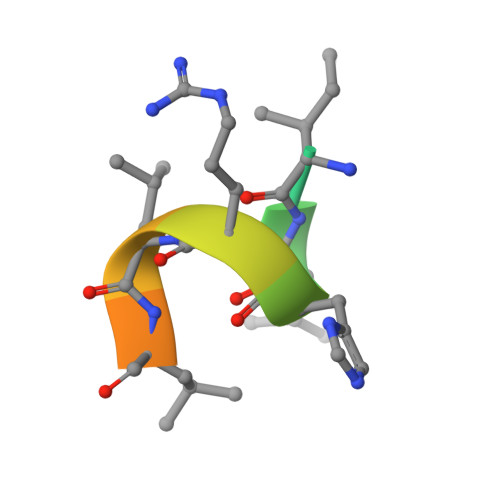
Estrogen receptor alpha somatic mutations Y537S and D538G confer breast cancer endocrine resistance by stabilizing the activating function-2 binding conformation.
Fanning, S.W., Mayne, C.G., Dharmarajan, V., Carlson, K.E., Martin, T.A., Novick, S.J., Toy, W., Green, B., Panchamukhi, S., Katzenellenbogen, B.S., Tajkhorshid, E., Griffin, P.R., Shen, Y., Chandarlapaty, S., Katzenellenbogen, J.A., Greene, G.L.(2016) Elife 5
- PubMed: 26836308
- DOI: https://doi.org/10.7554/eLife.12792
- Primary Citation of Related Structures:
4PXM, 4Q13, 4Q50 - PubMed Abstract:
Somatic mutations in the estrogen receptor alpha (ERα) gene (ESR1), especially Y537S and D538G, have been linked to acquired resistance to endocrine therapies. Cell-based studies demonstrated that these mutants confer ERα constitutive activity and antiestrogen resistance and suggest that ligand-binding domain dysfunction leads to endocrine therapy resistance. Here, we integrate biophysical and structural biology data to reveal how these mutations lead to a constitutively active and antiestrogen-resistant ERα. We show that these mutant ERs recruit coactivator in the absence of hormone while their affinities for estrogen agonist (estradiol) and antagonist (4-hydroxytamoxifen) are reduced. Further, they confer antiestrogen resistance by altering the conformational dynamics of the loop connecting Helix 11 and Helix 12 in the ligand-binding domain of ERα, which leads to a stabilized agonist state and an altered antagonist state that resists inhibition.
- Ben May Department for Cancer Research, University of Chicago, Chicago, United States.
Organizational Affiliation: