Discrete conformations of epitope II on the hepatitis C virus E2 protein for antibody-mediated neutralization and nonneutralization.
Deng, L., Ma, L., Virata-Theimer, M.L., Zhong, L., Yan, H., Zhao, Z., Struble, E., Feinstone, S., Alter, H., Zhang, P.(2014) Proc Natl Acad Sci U S A 111: 10690-10695
- PubMed: 25002515
- DOI: https://doi.org/10.1073/pnas.1411317111
- Primary Citation of Related Structures:
4Q0X - PubMed Abstract:
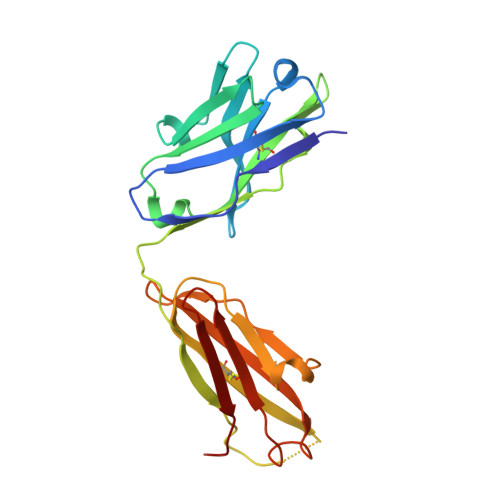
The X-ray crystal structure of epitope II on the E2 protein of hepatitis C virus, in complex with nonneutralizing antibody mAb#12, has been solved at 2.90-Å resolution. The spatial arrangement of the essential components of epitope II (ie, the C-terminal α-helix and the N-terminal loop) was found to deviate significantly from that observed in those corresponding complexes with neutralizing antibodies. The distinct conformations are mediated largely by the flexibility of a highly conserved glycine residue that connects these components. Thus, it is the particular tertiary structure of epitope II, which is presented in a spatial and temporal manner, that determines the specificity of antibody recognition and, consequently, the outcome of neutralization or nonneutralization.
- Division of Hematology, Office of Blood Research and Review, Center for Biologics Evaluation and Research, US Food and Drug Administration, Bethesda, MD 20892;
Organizational Affiliation: