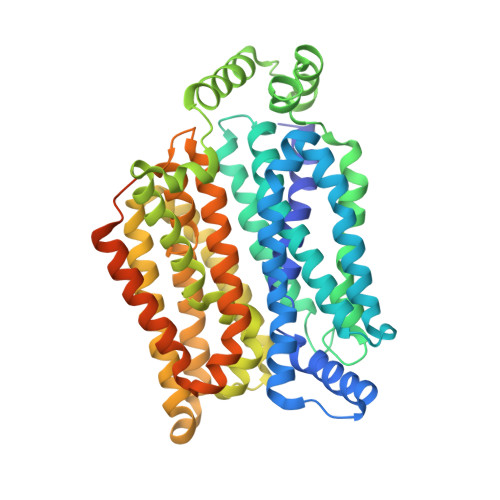
Crystal structure of the human glucose transporter GLUT1
Deng, D., Xu, C., Sun, P.C., Wu, J.P., Yan, C.Y., Hu, M.X., Yan, N.(2014) Nature 510: 121-125
- PubMed: 24847886
- DOI: https://doi.org/10.1038/nature13306
- Primary Citation of Related Structures:
4PYP - PubMed Abstract:
The glucose transporter GLUT1 catalyses facilitative diffusion of glucose into erythrocytes and is responsible for glucose supply to the brain and other organs. Dysfunctional mutations may lead to GLUT1 deficiency syndrome, whereas overexpression of GLUT1 is a prognostic indicator for cancer. Despite decades of investigation, the structure of GLUT1 remains unknown. Here we report the crystal structure of human GLUT1 at 3.2 Å resolution. The full-length protein, which has a canonical major facilitator superfamily fold, is captured in an inward-open conformation. This structure allows accurate mapping and potential mechanistic interpretation of disease-associated mutations in GLUT1. Structure-based analysis of these mutations provides an insight into the alternating access mechanism of GLUT1 and other members of the sugar porter subfamily. Structural comparison of the uniporter GLUT1 with its bacterial homologue XylE, a proton-coupled xylose symporter, allows examination of the transport mechanisms of both passive facilitators and active transporters.
- 1] State Key Laboratory of Bio-membrane and Membrane Biotechnology, Tsinghua University, Beijing 100084, China [2] Center for Structural Biology, School of Life Sciences and School of Medicine, Tsinghua University, Beijing 100084, China [3] Tsinghua-Peking Center for Life Sciences, Tsinghua University, Beijing 100084, China [4].
Organizational Affiliation: