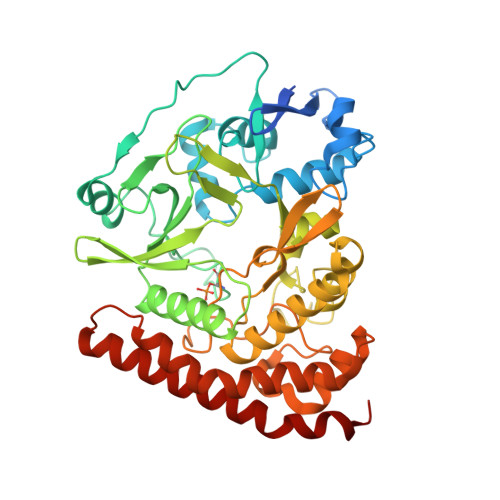
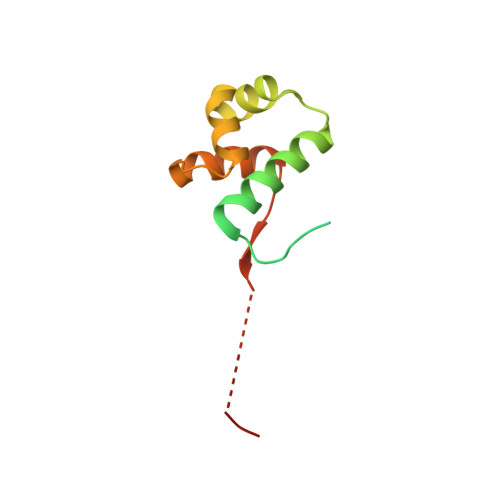
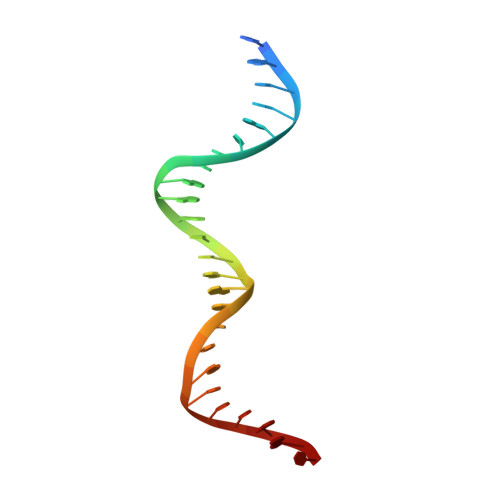
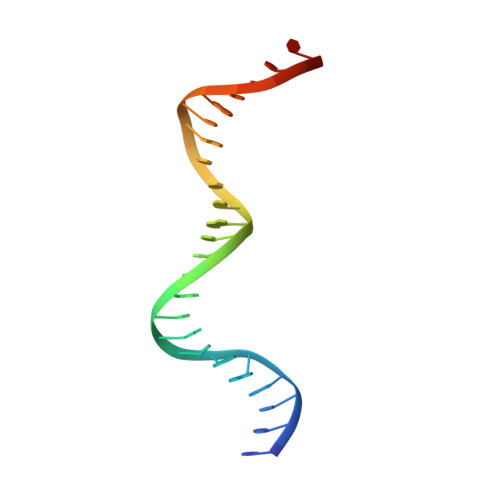
The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence.
Wen, Y., Behiels, E., Felix, J., Elegheert, J., Vergauwen, B., Devreese, B., Savvides, S.N.(2014) Nucleic Acids Res 42: 10134-10147
- PubMed: 25056321
- DOI: https://doi.org/10.1093/nar/gku665
- Primary Citation of Related Structures:
4PU3, 4PU4, 4PU5, 4PU7, 4PU8 - PubMed Abstract:
Nearly all bacteria exhibit a type of phenotypic growth described as persistence that is thought to underlie antibiotic tolerance and recalcitrant chronic infections. The chromosomally encoded high-persistence (Hip) toxin-antitoxin proteins HipASO and HipBSO from Shewanella oneidensis, a proteobacterium with unusual respiratory capacities, constitute a type II toxin-antitoxin protein module. Here we show that phosphorylated HipASO can engage in an unexpected ternary complex with HipBSO and double-stranded operator DNA that is distinct from the prototypical counterpart complex from Escherichia coli. The structure of HipBSO in complex with operator DNA reveals a flexible C-terminus that is sequestered by HipASO in the ternary complex, indicative of its role in binding HipASO to abolish its function in persistence. The structure of HipASO in complex with a non-hydrolyzable ATP analogue shows that HipASO autophosphorylation is coupled to an unusual conformational change of its phosphorylation loop. However, HipASO is unable to phosphorylate the translation factor Elongation factor Tu, contrary to previous reports, but in agreement with more recent findings. Our studies suggest that the phosphorylation state of HipA is an important factor in persistence and that the structural and mechanistic diversity of HipAB modules as regulatory factors in bacterial persistence is broader than previously thought.
- Unit for Biological Mass Spectrometry and Proteomics, Laboratory for Protein Biochemistry and Biomolecular Engineering (L-ProBE), Ghent University, K.L. Ledeganckstraat 35, 9000 Ghent, Belgium Unit for Structural Biology, Laboratory for Protein Biochemistry and Biomolecular Engineering (L-ProBE), Ghent University, K.L. Ledeganckstraat 35, 9000 Ghent, Belgium.
Organizational Affiliation: