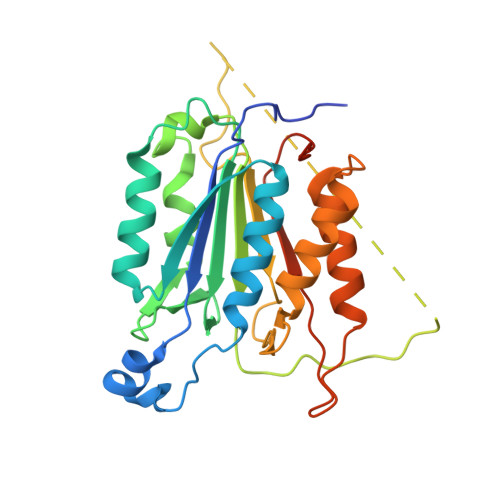
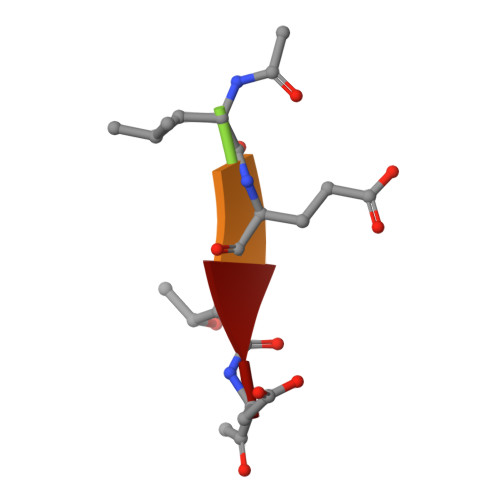
Selective inhibition of initiator versus executioner caspases using small peptides containing unnatural amino acids.
Vickers, C.J., Gonzalez-Paez, G.E., Litwin, K.M., Umotoy, J.C., Coutsias, E.A., Wolan, D.W.(2014) ACS Chem Biol 9: 2194-2198
- PubMed: 25079698
- DOI: https://doi.org/10.1021/cb5004256
- Primary Citation of Related Structures:
4PRY, 4PRZ, 4PS0, 4PS1 - PubMed Abstract:
Caspases are fundamental to many essential biological processes, including apoptosis, differentiation, and inflammation. Unregulated caspase activity is also implicated in the development and progression of several diseases, such as cancer, neurodegenerative disorders, and sepsis. Unfortunately, it is difficult to determine exactly which caspase(s) of the 11 isoforms that humans express is responsible for specific biological functions. This lack of resolution is primarily due to highly homologous active sites and overlapping substrates. Currently available peptide-based inhibitors and probes are based on specificity garnered from peptide substrate libraries. For example, the canonical tetrapeptide LETD was discovered as the canonical sequence that is optimally recognized by caspase-8; however, LETD-based inhibitors and substrates promiscuously bind to other isoforms with equal affinity, including caspases-3, -6, and -9. In order to mitigate this problem, we report the identification of a new series of compounds that are >100-fold selective for inhibiting the initiator caspases-8 and -9 over the executioner caspases-3, -6, and -7.
- Departments of Molecular and Experimental Medicine and Chemical Physiology, The Scripps Research Institute , La Jolla, California 92037, United States.
Organizational Affiliation: