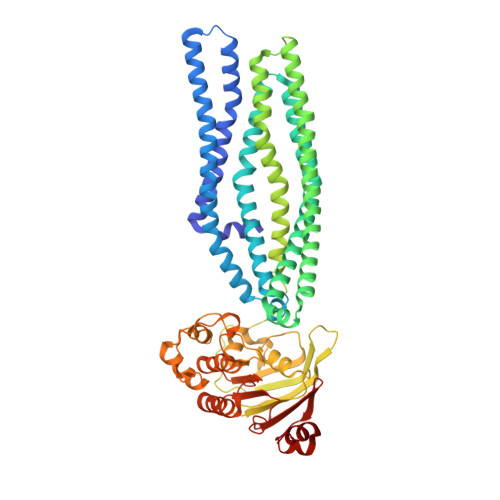
Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state.
Choudhury, H.G., Tong, Z., Mathavan, I., Li, Y., Iwata, S., Zirah, S., Rebuffat, S., van Veen, H.W., Beis, K.(2014) Proc Natl Acad Sci U S A 111: 9145
- PubMed: 24920594
- DOI: https://doi.org/10.1073/pnas.1320506111
- Primary Citation of Related Structures:
4PL0 - PubMed Abstract:
Enterobacteriaceae produce antimicrobial peptides for survival under nutrient starvation. Microcin J25 (MccJ25) is an antimicrobial peptide with a unique lasso topology. It is secreted by the ATP-binding cassette (ABC) exporter McjD, which ensures self-immunity of the producing strain through efficient export of the toxic mature peptide from the cell. Here we have determined the crystal structure of McjD from Escherichia coli at 2.7-Å resolution, which is to the authors' knowledge the first structure of an antibacterial peptide ABC transporter. Our functional and biochemical analyses demonstrate McjD-dependent immunity to MccJ25 through efflux of the peptide. McjD can directly bind MccJ25 and displays a basal ATPase activity that is stimulated by MccJ25 in both detergent solution and proteoliposomes. McjD adopts a new conformation, termed nucleotide-bound outward occluded. The new conformation defines a clear cavity; mutagenesis and ligand binding studies of the cavity have identified Phe86, Asn134, and Asn302 as important for recognition of MccJ25. Comparisons with the inward-open MsbA and outward-open Sav1866 structures show that McjD has structural similarities with both states without the intertwining of transmembrane (TM) helices. The occluded state is formed by rotation of TMs 1 and 2 toward the equivalent TMs of the opposite monomer, unlike Sav1866 where they intertwine with TMs 3-6 of the opposite monomer. Cysteine cross-linking studies on the McjD dimer in inside-out membrane vesicles of E. coli confirmed the presence of the occluded state. We therefore propose that the outward-occluded state represents a transition intermediate between the outward-open and inward-open conformation of ABC exporters.
- Division of Molecular Biosciences, Imperial College London, London SW7 2AZ, United Kingdom;Membrane Protein Lab, Diamond Light Source, Harwell Science and Innovation Campus, Chilton OX11 0DE, United Kingdom;Rutherford Appleton Laboratory, Research Complex at Harwell, Didcot OX11 0FA, United Kingdom;
Organizational Affiliation: