Crystal structure of L-methionine gamma-lyase from Citrobacter freundii with Y58F substitution
Revtovich, S.V., Nikulin, A.D., Anufrieva, N.V., Morozova, E.A., Demidkina, T.V.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| METHIONINE GAMMA-LYASE | 396 | Citrobacter freundii GTC 09479 | Mutation(s): 1 Gene Names: H262_14272 | 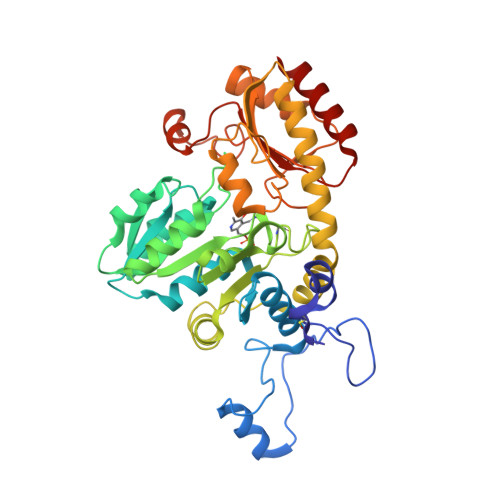 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 1PE Query on 1PE | C [auth A] | PENTAETHYLENE GLYCOL C10 H22 O6 JLFNLZLINWHATN-UHFFFAOYSA-N |  | ||
| PEG Query on PEG | B [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| Modified Residues 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CSO Query on CSO | A | L-PEPTIDE LINKING | C3 H7 N O3 S |  | CYS |
| LLP Query on LLP | A | L-PEPTIDE LINKING | C14 H22 N3 O7 P |  | LYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 57.546 | α = 90 |
| b = 124.531 | β = 90 |
| c = 129.874 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Russian Foundation for Basci Research | Russian Federation | -- |