Structural Conservation, Variability, and Immunogenicity of the T6 Backbone Pilin of Serotype M6 Streptococcus pyogenes.
Young, P.G., Moreland, N.J., Loh, J.M., Bell, A., Atatoa Carr, P., Proft, T., Baker, E.N.(2014) Infect Immun 82: 2949-2957
- PubMed: 24778112
- DOI: https://doi.org/10.1128/IAI.01706-14
- Primary Citation of Related Structures:
4P0D - PubMed Abstract:
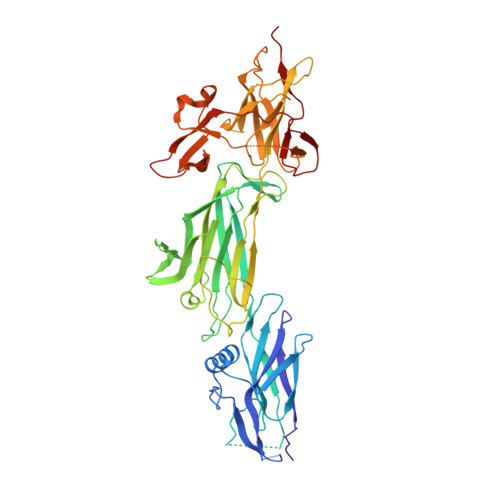
Group A streptococcus (GAS; Streptococcus pyogenes) is a Gram-positive human pathogen that causes a broad range of diseases ranging from acute pharyngitis to the poststreptococcal sequelae of acute rheumatic fever. GAS pili are highly diverse, long protein polymers that extend from the cell surface. They have multiple roles in infection and are promising candidates for vaccine development. This study describes the structure of the T6 backbone pilin (BP; Lancefield T-antigen) from the important M6 serotype. The structure reveals a modular arrangement of three tandem immunoglobulin-like domains, two with internal isopeptide bonds. The T6 pilin lysine, essential for polymerization, is located in a novel VAKS motif that is structurally homologous to the canonical YPKN pilin lysine in other three- and four-domain Gram-positive pilins. The T6 structure also highlights a conserved pilin core whose surface is decorated with highly variable loops and extensions. Comparison to other Gram-positive BPs shows that many of the largest variable extensions are found in conserved locations. Studies with sera from patients diagnosed with GAS-associated acute rheumatic fever showed that each of the three T6 domains, and the largest of the variable extensions (V8), are targeted by IgG during infection in vivo. Although the GAS BP show large variations in size and sequence, the modular nature of the pilus proteins revealed by the T6 structure may aid the future design of a pilus-based vaccine.
- School of Biological Sciences, University of Auckland, Auckland, New Zealand Maurice Wilkins Centre, University of Auckland, Auckland, New Zealand p.young@auckland.ac.nz.
Organizational Affiliation: