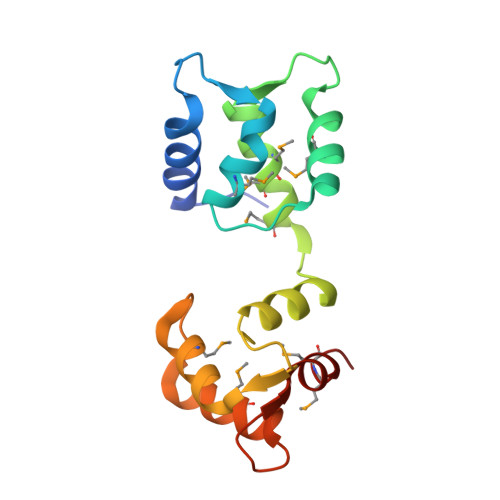
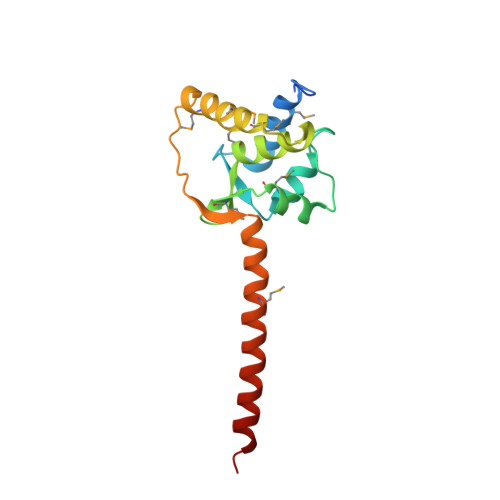
Regulation of the NaV1.5 cytoplasmic domain by calmodulin.
Gabelli, S.B., Boto, A., Kuhns, V.H., Bianchet, M.A., Farinelli, F., Aripirala, S., Yoder, J., Jakoncic, J., Tomaselli, G.F., Amzel, L.M.(2014) Nat Commun 5: 5126
- PubMed: 25370050
- DOI: https://doi.org/10.1038/ncomms6126
- Primary Citation of Related Structures:
4OVN - PubMed Abstract:
Voltage-gated sodium channels (Na(v)) underlie the rapid upstroke of action potentials in excitable tissues. Binding of channel-interactive proteins is essential for controlling fast and long-term inactivation. In the structure of the complex of the carboxy-terminal portion of Na(v)1.5 (CTNa(v)1.5) with calmodulin (CaM)-Mg(2+) reported here, both CaM lobes interact with the CTNa(v)1.5. On the basis of the differences between this structure and that of an inactivated complex, we propose that the structure reported here represents a non-inactivated state of the CTNa(v), that is, the state that is poised for activation. Electrophysiological characterization of mutants further supports the importance of the interactions identified in the structure. Isothermal titration calorimetry experiments show that CaM binds to CTNa(v)1.5 with high affinity. The results of this study provide unique insights into the physiological activation and the pathophysiology of Na(v) channels.
- 1] Structural Enzymology and Thermodynamics Group, Department of Biophysics and Biophysical Chemistry, Johns Hopkins University School of Medicine, 725 N Wolfe Street, WBSB 608, Baltimore, Maryland 21205, USA [2] Division of Cardiology, Department of Medicine, Johns Hopkins University School of Medicine, 720 Rutland Avenue, Ross Building 844, Baltimore, Maryland 21205, USA [3] Department of Oncology, Johns Hopkins University School of Medicine, Baltimore, Maryland 21287, USA.
Organizational Affiliation: