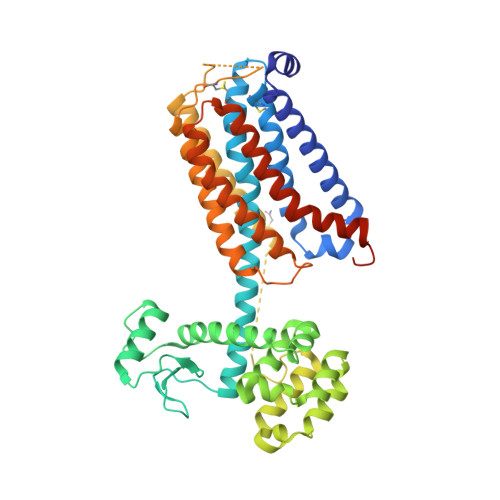
Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain.
Dore, A.S., Okrasa, K., Patel, J.C., Serrano-Vega, M., Bennett, K., Cooke, R.M., Errey, J.C., Jazayeri, A., Khan, S., Tehan, B., Weir, M., Wiggin, G.R., Marshall, F.H.(2014) Nature 511: 557-562
- PubMed: 25042998
- DOI: https://doi.org/10.1038/nature13396
- Primary Citation of Related Structures:
4OO9 - PubMed Abstract:
Metabotropic glutamate receptors are class C G-protein-coupled receptors which respond to the neurotransmitter glutamate. Structural studies have been restricted to the amino-terminal extracellular domain, providing little understanding of the membrane-spanning signal transduction domain. Metabotropic glutamate receptor 5 is of considerable interest as a drug target in the treatment of fragile X syndrome, autism, depression, anxiety, addiction and movement disorders. Here we report the crystal structure of the transmembrane domain of the human receptor in complex with the negative allosteric modulator, mavoglurant. The structure provides detailed insight into the architecture of the transmembrane domain of class C receptors including the precise location of the allosteric binding site within the transmembrane domain and key micro-switches which regulate receptor signalling. This structure also provides a model for all class C G-protein-coupled receptors and may aid in the design of new small-molecule drugs for the treatment of brain disorders.
- 1] Heptares Therapeutics Ltd, BioPark, Broadwater Road, Welwyn Garden City, Hertfordshire AL7 3AX, UK [2].
Organizational Affiliation: