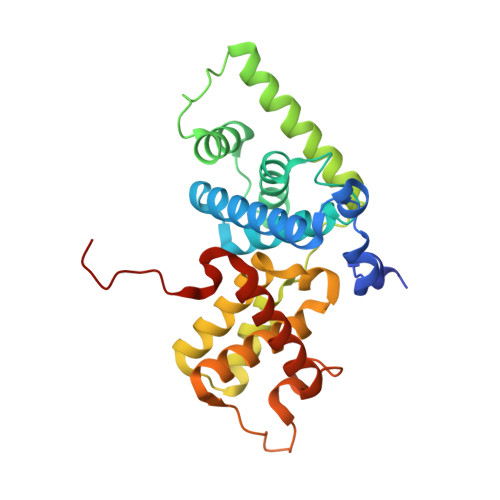
AFF4 binding to Tat-P-TEFb indirectly stimulates TAR recognition of super elongation complexes at the HIV promoter.
Schulze-Gahmen, U., Lu, H., Zhou, Q., Alber, T.(2014) Elife 3: e02375-e02375
- PubMed: 24843025
- DOI: https://doi.org/10.7554/eLife.02375
- Primary Citation of Related Structures:
4OGR - PubMed Abstract:
Superelongation complexes (SECs) are essential for transcription elongation of many human genes, including the integrated HIV-1 genome. At the HIV-1 promoter, the viral Tat protein binds simultaneously to the nascent TAR RNA and the CycT1 subunit of the P-TEFb kinase in a SEC. To understand the preferential recruitment of SECs by Tat and TAR, we determined the crystal structure of a quaternary complex containing Tat, P-TEFb, and the SEC scaffold, AFF4. Tat and AFF4 fold on the surface of CycT1 and interact directly. Interface mutations in the AFF4 homolog AFF1 reduced Tat-AFF1 affinity in vivo and Tat-dependent transcription from the HIV promoter. AFF4 binding in the presence of Tat partially orders the CycT1 Tat-TAR recognition motif and increases the affinity of Tat-P-TEFb for TAR 30-fold. These studies indicate that AFF4 acts as a two-step filter to increase the selectivity of Tat and TAR for SECs over P-TEFb alone.DOI: http://dx.doi.org/10.7554/eLife.02375.001.
- Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, United States uschulze-gahmen@berkeley.edu.
Organizational Affiliation: