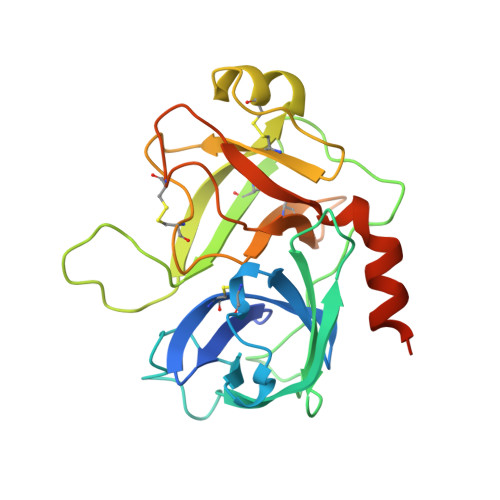
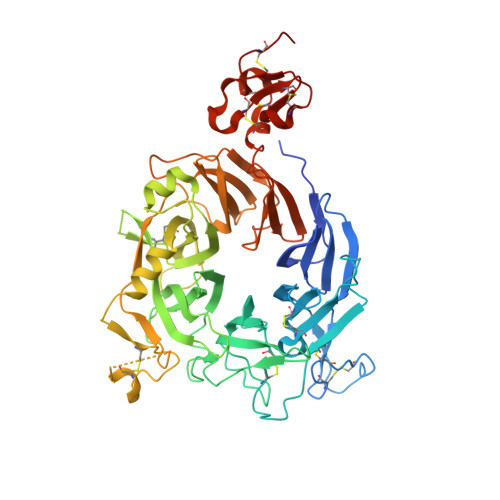
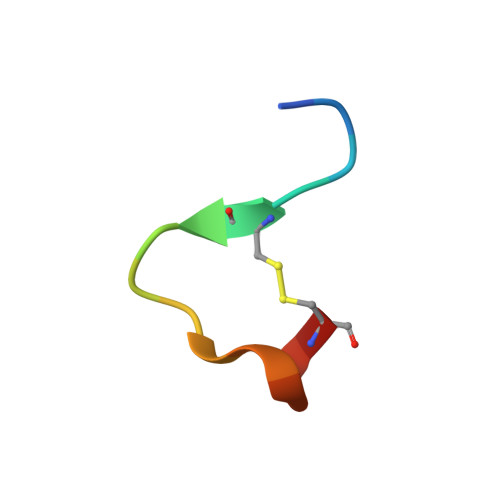
An allosteric switch for pro-HGF/Met signaling using zymogen activator peptides.
Landgraf, K.E., Steffek, M., Quan, C., Tom, J., Yu, C., Santell, L., Maun, H.R., Eigenbrot, C., Lazarus, R.A.(2014) Nat Chem Biol 10: 567-573
- PubMed: 24859116
- DOI: https://doi.org/10.1038/nchembio.1533
- Primary Citation of Related Structures:
4O3T, 4O3U - PubMed Abstract:
Stimulation of hepatocyte growth factor (HGF) signaling through the Met receptor is an attractive approach for promoting tissue repair and preventing fibrosis. Using structure-guided peptide phage display combined with an activity-based sorting strategy, we engineered allosteric activators of zymogen-like pro-HGF to bypass proteolytic activation and reversibly stimulate pro-HGF signaling through Met. Biochemical, structural and biological data showed that zymogen activator peptides (ZAPtides) potently and selectively bind the activation pocket within the serine protease-like β-chain of pro-HGF and display titratable activation of pro-HGF-dependent Met signaling, leading to cell survival and migration. To further demonstrate the versatility of our ZAPtide platform, we identified allosteric activators for pro-macrophage stimulating protein and a zymogen serine protease, Protein C, which also provides evidence for target selectivity. These studies reveal that ZAPtides use molecular mimicry of the trypsin-like N-terminal insertion mechanism and establish a new paradigm for selective pharmacological activation of plasminogen-related growth factors and zymogen serine proteases.
- 1] Department of Early Discovery Biochemistry, Genentech, Inc., San Francisco, California, USA. [2].
Organizational Affiliation: